18 Recommended R server specs
We conducted some stress tests to assess the maximum allocated RAM and HDD space consumed by the analysis portraited on this bookdown.
18.1 Limma + voom
For epigenomics and differential gene expression analysis we portrayed the use of limma + voom. The disk space consumed by the ExpressionSets used is dependent on the array dimensions and the number of individuals, and so is the RAM allocation; this yields the following table:
| Array | Individuals | Storage [GB] | RAM [GB] |
|---|---|---|---|
| EPIC 450K | 100 | 0,34 | 3 |
| EPIC 450K | 500 | 1,7 | 12 |
| EPIC 450K | 1000 | 3,4 | 24 |
| EPIC 450K | 2000 | 6,8 | 48 |
| EPIC 850K | 100 | 0,64 | 8 |
| EPIC 850K | 500 | 3,2 | 36 |
| EPIC 850K | 1000 | 6,4 | 72 |
| EPIC 850K | 2000 | 12,8 | 144 |
This table can be summarized on the following graphs:
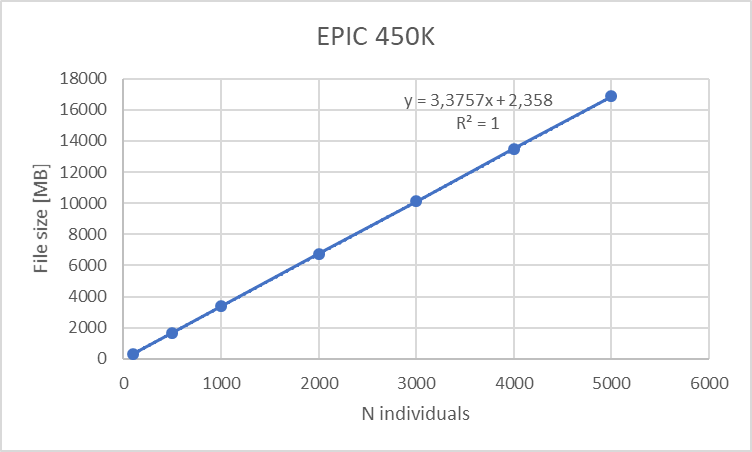
Figure 18.1: Disk space requirements for EPIC 450K array versus number of individuals (10 meta-variables)
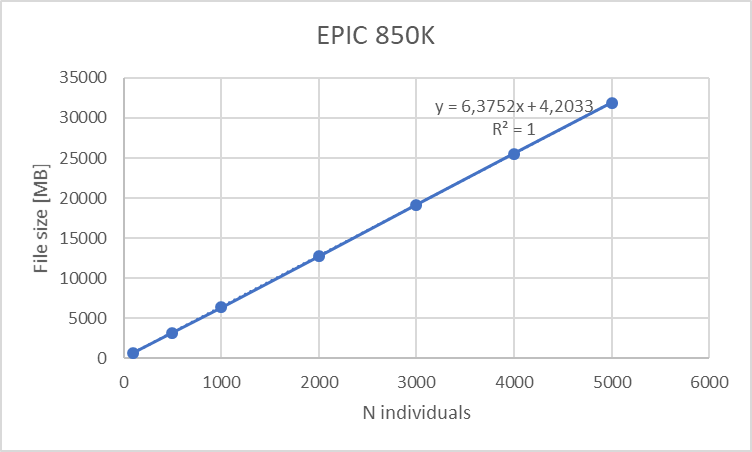
Figure 18.2: Disk space requirements for EPIC 850K array versus number of individuals (10 meta-variables)
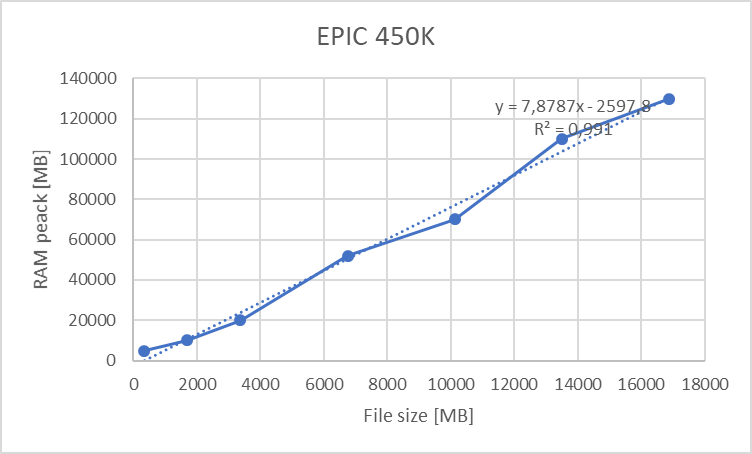
Figure 18.3: Maximum allocated RAM for EPIC 450K array versus number of individuals (1 adjustin covariable)
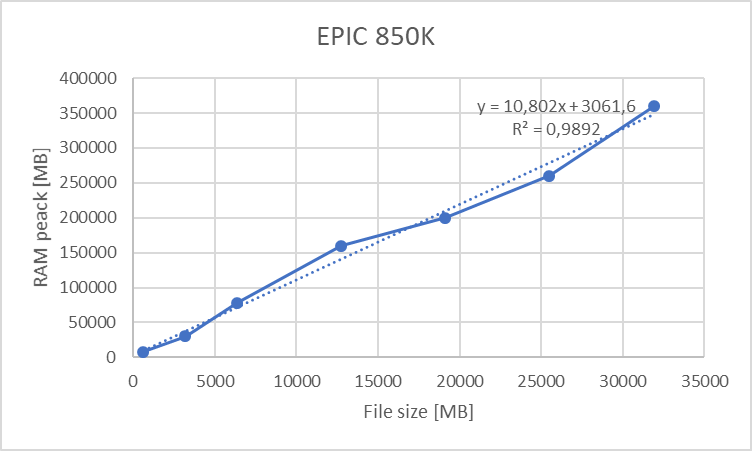
Figure 18.4: Maximum allocated RAM for EPIC 850K array versus number of individuals (1 adjustin covariable)
We can clearly see a lineal correlation to estimate the hardware requirements.
18.2 GWAS + PCA
Genomic data is accepted in OmicSHIELD as gds files, which can be compressed using different algorithms, therefore it makes no sense to extract any relationship between individuals / number of variants and file size. Please refer to the appropiate section for information about that.
Also, the methodologies used (both pooled and meta-analysis) are not as RAM dependent as Limma analysis. This has to do with the nature of genomic data, which is typically extremely large; to analyze this kind of data we do not load it entirely on the RAM, the algorithms perform clever techniques that read portions of the data at a time, and the size of that portion can be configured by the user (see the argument snpBlock), therefore the researchers can tune the function calls if RAM limits become an issue.
18.3 General remarks
On top of this information some details must be noted:
- Disk space (storage) is cheap and easy to upgrade, RAM is expensive. Therefore storage requirements should be easy to fulfill and RAM requirements may require a steeper investment.
- The server has to perform the analysis AND host the Opal/Armadillo + R server. For reference, our deployment, which contains Apache + Opal + 4 x R servers, allocates 5 GB of RAM memory.
- Results are approximate, it is advisable to apply a security coefficient (especially on the RAM) of about 1.5 to be safe.
- Imputed GWAS data can be quite heavy on disk space, however it is not that heavy on RAM usage (https://www.well.ox.ac.uk/~gav/snptest/#other_options chunk flag).
- Methylation data in contrast is more RAM intensive when being analyzed.
- There is no queuing system implemented at the moment, so if multiple people are conducting analyses at the same time, the performance of the system will be compromised.
- If possible, get more RAM and storage than the bare minimum, as it is expected that omics data will increase in longitudinal cohort studies during next years.
Regarding the discussion of dimensioning the servers for number of cores; this does not depend on the size and complexity of the analysis to be carried out, but some functionalities benefit from a multi-core architecture to speed up the calculations. However, it is not a constraint, as everything will run perfectly on one core. The discussion to take place is about the number of concurrent users of the servers. As a rule of thumb, we recommend having one core per user, that way things will not collapse when all users perform analyses at the same time. However, this has financial consequences. A good starting point would be to work with 8 cores minimum, but this is for the partners to decide.