3 Datasets description
Here, we will describe the data employed for the analyses presented in the bookdown, giving details about experimental designs, the type of features available, and the structure and organization of data in the Opal infrastructure. Employed data constitute both public data downloaded from public repositories, and real datasets deriving from an European H2020 research project on exposome and health (HELIX Project). The public datasets have been uploaded to the Opal demo site and can be easily accessed without additional permissions. On the other hand, the HELIX dataset is available in the Opal BRGE site hosted by the Bioinformatic Research Group in Epidemiology of ISGlobal. In both cases, data are organized in the Opal simulating a single-site DataSHIELD infrastructure. Details for accessing the server will be found in each section, at the beginning of each pipeline.
3.1 CINECA project
Data from the public CINECA project were employed in this bookdown for demonstrating the capabilities of dsOmicSHIELD for performing GWAS and other genetic analyses. Data correspond to synthetic data set generated by the CINECA Project made freely available under the Creative Commons Licence (CC-BY) (funding: EC H2020 grant 825775). In Figure 3.1, an overall description of the datasets and cohorts belonging to the CINECA project is presented.
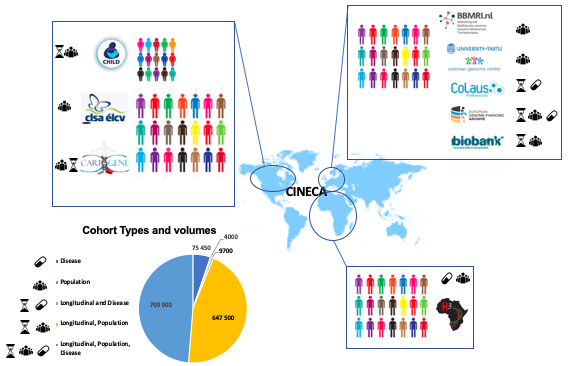
Figure 3.1: The figure depicts CINECA project cohorts representing >1.4 Million individuals and spanning three continents.
3.2 Data organization in the Opal server
Data were initially accessible from a public EGA repository (ID: EGAD00001006673), being a deliverable of the VEIS project. Previous to analysis, data were pruned, and the individuals were separated into three synthetic cohorts. Information on individuals and SNP count of each cohort can be found on the Table 3.1.
| Cohort 1 | Cohort 2 | Cohort 3 | Total | |
|---|---|---|---|---|
| Number of SNPs | 865,240 | 865,240 | 865,240 | 865,240 |
| Number of individuals | 817 | 1,073 | 614 | 2,504 |
The three datasets were uploaded to the Opal demo site organized as (1) an array of resource objects, corresponding to unitary VCF files with variant information (one per chromosome and cohort), and (2) table objects corresponding to the phenotype data (one per cohort). Data are in Opal simulating a single-site DataSHIELD architecture, as in Figure 3.2. In this figure, the data analyst corresponds to the “RStudio” session, which through the DataSHIELD Interface (DSI) connects with the Opal server located at the cohort network.
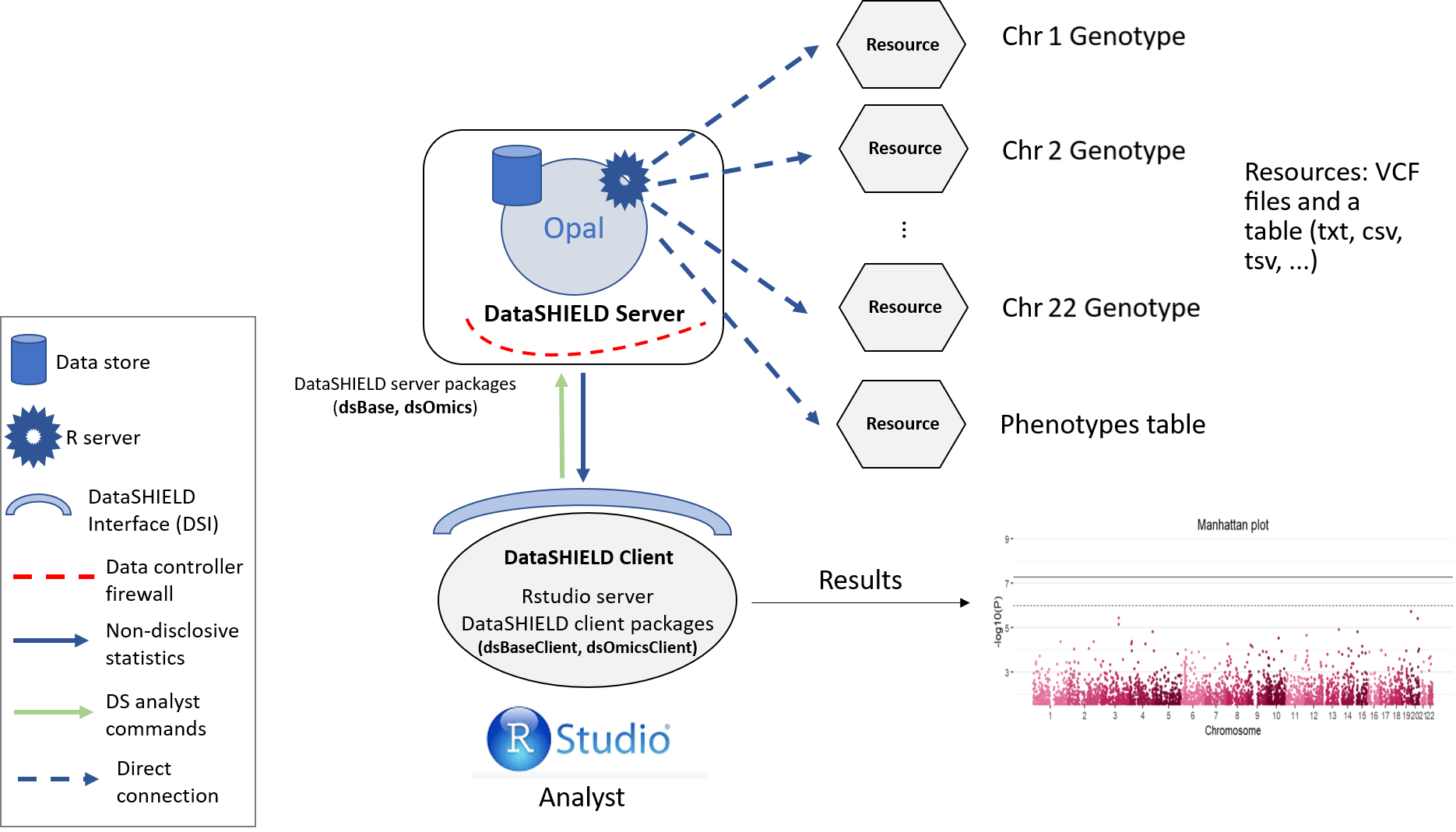
Figure 3.2: Proposed infrastructure to perform single-cohort GWAS studies.
For more information about DataSHIELD data formats, please, refers to the bookdown “Orchestrating privacy-protected non-disclosive big data analyses of data from different resources with R and DataSHIELD”.
3.3 TCGA Liver project
The public TCGA Liver project was selected for illustrating how to perform transcriptomic data analysis in DataSHIELD. These data derived from the famous TCGA project. We have uploaded to the demo Opal server a resource called tcga_liver whose URL is http://duffel.rail.bio/recount/TCGA/rse_gene_liver.Rdata available through the recount project.
Some summaries of the dataset are the following:
| TCGA Liver data | |
|---|---|
| Number of individuals | 424 |
| Number of genes | 58,037 |
| Number of covariate fields | 864 |
The RangedSummarizedExperiment (RSE) objects are part of the BioConductor project, as a brief summary, these types of objects are designed to contain different multi-omic data, they can also contain the individuals phenotypes and additional metadata associated with a particular experiment. For further information about what are RangedSummarizedExperiment please take a look at the official documentation
The resource used on this project contains a RangedSummarizedExperiment with the RNAseq profiling of liver cancer data from TCGA. The resource is located inside the OMICS project. The structure used is illustrated on the following figure.
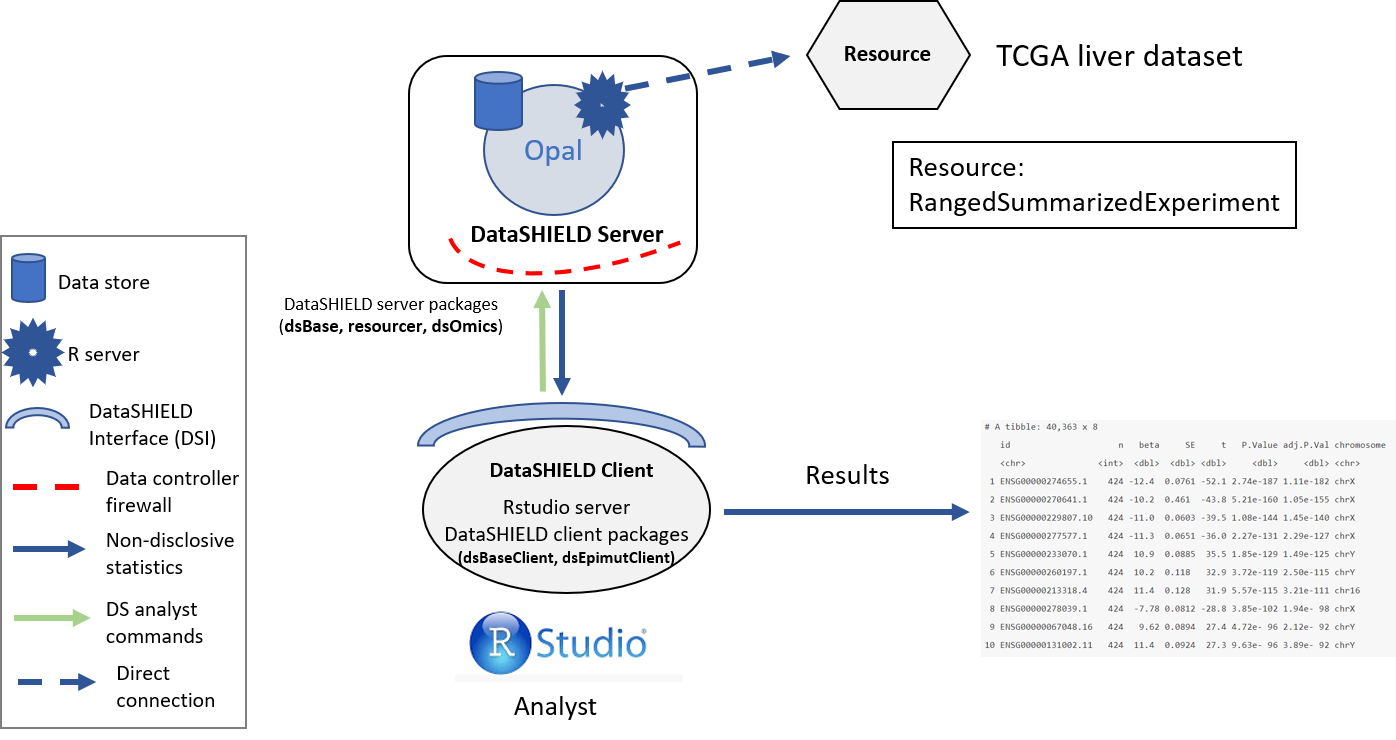
Figure 3.3: Proposed infrastructure to perform DGE studies.
The data analyst corresponds to the “RStudio” session, which through DataSHIELD Interface (DSI) connects with the Opal server. This Opal server contains a resource that corresponds to the TCGA liver RSE.
3.4 Epigenetic data from the GSE66351 project
The public data from the GSE66351 project were used in this section for illustrating how to perform EWAS in DataSHIELD. They has been downloaded from GEO (accession number GSE66351) and consist on DNA methylation reads from the Illumina 450K array. Data corresponds to CpGs beta values measured in the superior temporal gyrus and prefrontal cortex brain regions of patients with Alzheimer’s.
This kind of data is encapsulated on a type of R object called ExpressionSet, this objects are part of the BioConductor project and are meant to contain different sources of genomic data, alongside the genomic data they can also contain the phenotypes and metadata associated to a study. Researchers who are not familiar with ExpressionSet can find further information here. Notice that genomic data is encoded as beta-values that ensure data harmonization across studies.
In order to illustrate how to perform data analyses using federated data, we have split the data into two synthetic cohorts (split by individuals). We have created two resources on the demo Opal server called GSE66351_1 and GSE66351_2 respectively. They can be found inside the OMICS project. The structure used is illustrated on the following figure.
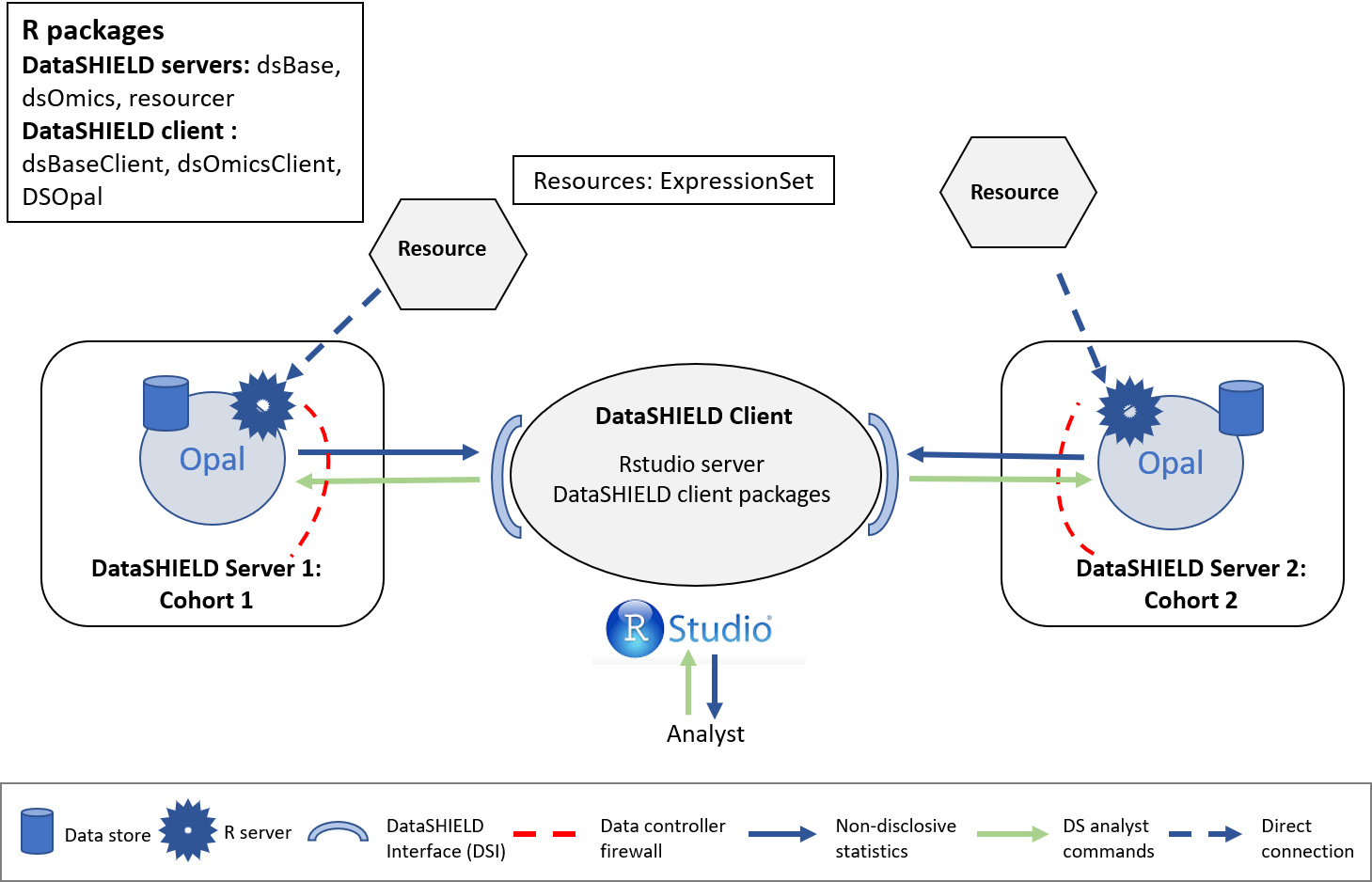
Figure 3.4: Proposed infrastructure to perform EWAS studies.
Some summaries of the datasets are the following:
| Cohort 1 | Cohort 2 | Total | |
|---|---|---|---|
| Number of CpGs | 481,868 | 481,868 | 481,868 |
| Number of individuals | 100 | 90 | 190 |
| Number of covariates | 49 | 49 | 49 |
| Number of annotation features | 37 | 37 | 37 |
3.5 HELIX project
The real-world research problem presented in this book comes from the HELIX Project. The HELIX (Human Early-Life Exposome) project gathers data from 6 longitudinal-based European birth cohorts, with the aim of evaluating the effect of environmental risk factors on mothers and children health, with a special focus in the effects on molecular health profiles (omic data). HELIX cohorts include the BIB (Born in Bradford) (United Kingdom), EDEN (Étude des Déterminants pré et postnatals du développement et de la santé de l’ENfant) (France), INMA (INfancia y Medio Ambiente) (Spain), KANC (Kaunus Cohort) (Lithuania), MoBa (Norwegian Mother and Child Cohort Study) (Norway), and Rhea (Mother-Child Cohort in Crete) (Greece). General details of the study design of HELIX can be found in the Figure 3.5. The whole HELIX dataset includes a total of 31,472 mother-child pairs. Among them, a subcohort of 1,301 children (approximately 200 children in each cohort) was selected here according to the following criteria of eligibility: 1) age 6 to 11 years; 2) omic data available (including gene expression and EWAS data); 3) complete address history; and 4) no serious health problems that may affect the clinical testing or the child safety.

Figure 3.5: General overview of the HELIX research project.
In the 1,301 children, a wide range of environmental exposures were evaluated to define the early-life exposome during two time periods: the prenatal pregnancy period and childhood (age 6 to 11 years). Collected exposures can be organized into three main parts of the exposome: outdoor exposures, chemical exposures measured through biomarkers, and lifestyle factors. All variables incorporated in the dataset have been appropriately pre-processed previous to analysis (outliers removal, normalization, and missing values imputation). Regarding phenotype data, a wide range of health outcomes are also available for the childhood period including phenotypes related to (1) obesity and cardiometabolic health, (2) respiratory health, and (3) cognition and mental health. Finally, omic data (mainly transcriptomic and genome-wide DNA methylation data) are also available in the childhood period. The objective of this dataset was illustrating a real case study of:
- Differential gene expression analysis.
- Differential DNA methylation analysis.
3.5.1 Data organization in the Opal server
Omic data from the HELIX project were uploaded into to the Opal BRGE site simulating a single-site DataSHIELD architecture, as illustrated in Figure 3.6. Data were stored under the form of resources (one per cohort and data type). As illustrated in the figure, transcriptomic data) were stored under the form of expressionSets while EWAS data were stored as GenomicRatioSet. A summary of the structure and organization of this data in the Opal for the HELIX project is illustrated in Figure 3.6.
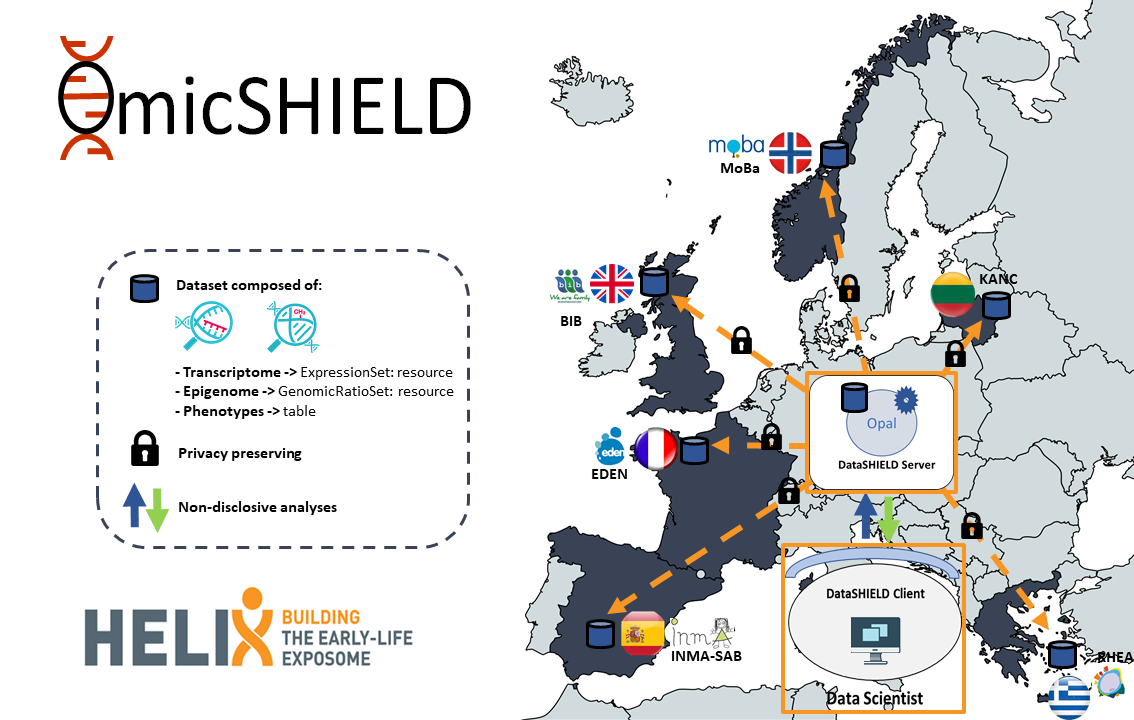
Figure 3.6: Input HELIX datasets organized by cohort in the Opal server.
In the next subsection, we present extra details for each of these datasets.
3.5.1.1 Gene expression data
The gene expression data available for the HELIX cohorts correspond to microarray data deriving from the Human Transcriptome Array 2.0 platform of Affymetrix. These are hosted in the the Opal BRGE site as ExpressionSet objects. Particularly, one ExpressionSet file per cohort is hosted in the Opal BRGE site in the form of a resource, as presented in the Figure 3.7. As it can be seen, data are organized within the Opal server in a project called HELIX_omics.
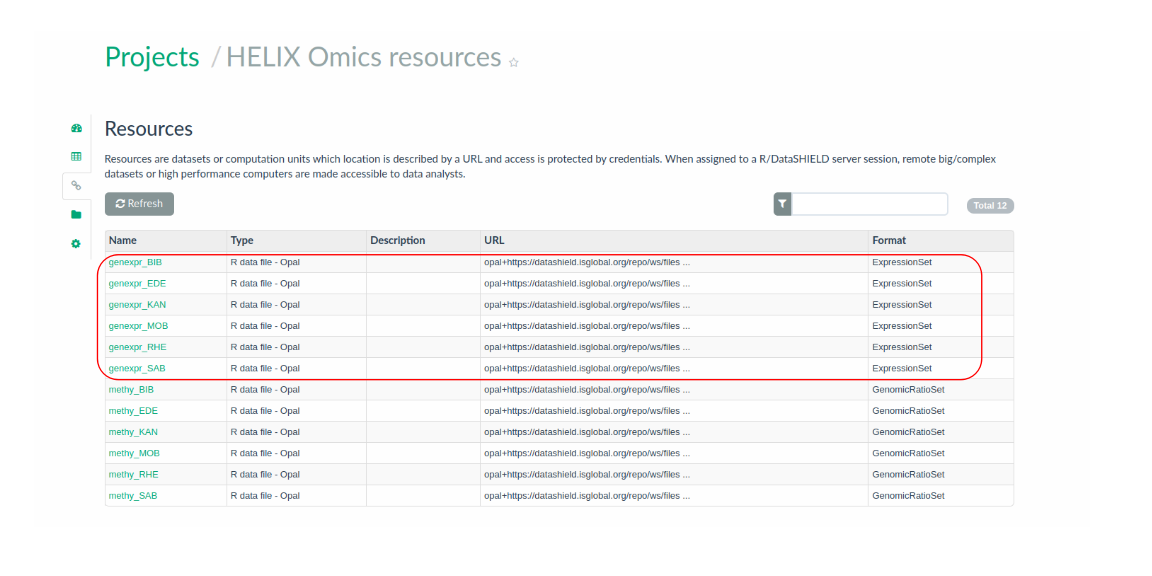
Figure 3.7: Input ExposomeSet data organized by cohort in the Opal server as resources.
3.5.1.2 Epigenetic data
The epigenetic data available for the HELIX cohorts correspond to genome-wide DNA methylation data obtained by microarray experiments using the Infinium HumanMethylation450k platform from Illumina. The data format in which these data are hosted in the Opal is the type GenomicRatioSet. GenomicRatioSet is a extend of the class SummarizedExperiment especially designed for DNA methylation data obtained with the Infinium HumanMethylation450k platform, that will be analysed with the associated R package minfi. This type of data usually contain pre-processed DNA methylation values at a genome-wide level, in the form of M or/and Beta values, together with the associated genomic coordinates. As in the case of ExpressionSet types, GenomicRatioSet can also incorporate phenotype and metadata information.
Particularly, one GenomicRatioSet file per cohort is hosted in the Opal BRGE site in the form of a resource, as presented in the Figure 3.8. As it can be seen, data are organized within the Opal server in a project called HELIX_omics.
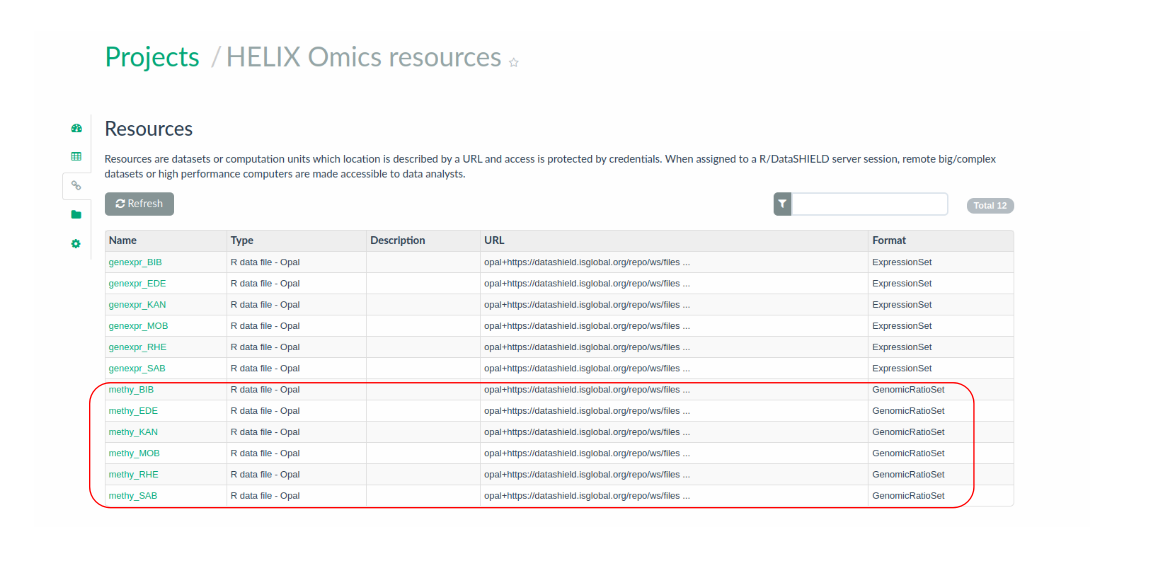
Figure 3.8: Input GenomicRatioSet data organized by cohort in the Opal server as resources.