11 Transcriptomic analysis in the HELIX cohort.
In this section, we will illustrate how to perform gene expression analysis using data from the HELIX project. Particularly, we will analyse microarray data deriving from the Human Transcriptome Array 2.0 Affymetrix platform. On these data, we will show how to perform a differential gene expression analysis comparing the transcriptomic profiles of boys and girls in the HELIX cohorts.
The microarray data to be analysed has been previously pre-processed (e.g., normalized, low expression or cross-reactive probes removed, etc.), so, this showcase will be restricted to the analysis part. For that, we will employ the dsOmicsClient functions that adapt some of the most important “limma” methods for their use in dataSHIELD. Please, note that pre-processing of gene expression data is also included in dsOmicsClient as indicated in our bookdown. For this tutorial, data are hosted in the Opal BRGE site simulating a single-site DataSHIELD architecture, as previously described in the section 3.5.1.1.
11.1 Getting started.
In this section, we will describe how to configure R and DataSHIELD with the needed packages to carry out proposed analyses in remote. We start by installing the client-side version of the following DataSHIELD/Opal integration packages.
install.packages("DSOpal", dependencies=TRUE)
install.packages("DSI", dependencies=TRUE)Make sure you also install the DataSHIELD client-side version of the package dsBaseClient.
install.packages("dsBaseClient",
repos = c("http://cran.datashield.org","https://cloud.r-project.org/"),
dependencies = TRUE)Then, install the client-side version of the dsOmicsClient package directly from GitHub.
install.packages("devtools")
require("devtools")
devtools::install_github("isglobal-brge/dsOmicsClient",ref="master",force=TRUE)Once installations are completed, all the packages are loaded as usual.
require(DSOpal)
require(DSI)
require(dsBaseClient)
require(dsOmicsClient)
# Loading additional required packages (if not installed, you can easly install them using the BiocManager::install() function)
require(clusterProfiler)
require(org.Hs.eg.db)
require(ggplot2)
require(biomaRt)11.2 Data formatting and manipulation in DataSHIELD.
In this section, we will cover how to load and inspect input microarray data with DataSHIELD. We start by creating the connection to the opal server using an user who have DataSHIELD permissions.
builder <- DSI::newDSLoginBuilder()
builder$append(server = "BIB", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
builder$append(server = "EDEN", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
builder$append(server = "KANC", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
builder$append(server = "MoBA", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
builder$append(server = "Rhea", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
builder$append(server = "INMASAB", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
profile = "omics")
logindata <- builder$build()
conns <- DSI::datashield.login(logins = logindata)Now that we have created a connection object to the Opal, we have started a new R session on the server, and our analysis will take place in this remote session, so we have to load the data into it. In this use case, available data are in the form ExpressionSet files. ExpressionSet is a file-format of the BioConductor project that may contain different sources of genomic data. Alongside the genomic data, ExpressionSet may also contain some phenotypes and metadata associated to the population. Researchers who are not familiar with ExpressionSet can find further information here.
For our showcase, one ExpressionSet file per cohort is hosted in the Opal BRGE site in the form of a resource, all organized within the Opal server in a project called “OMICS”.
For loading these data into the remote DataSHIELD session we will use the DSI::datashield.assign.resource() function. This function takes the connections to the server created in the previous code chunk to assign all available resource objects from a project in the Opal to an R object in the DataSHIELD remote session. As it can be seen in the code, resources in DataSHIELD are called in the function following the next structure “NameOfOpalProject.NameOfResource”.
# We assign microarray data from all cohorts to an object called resource_pos
DSI::datashield.assign.resource(conns[1], "resource_expr",
"OMICS.genexpr_BIB")
DSI::datashield.assign.resource(conns[2], "resource_expr",
"OMICS.genexpr_EDE")
DSI::datashield.assign.resource(conns[3], "resource_expr",
"OMICS.genexpr_KAN")
DSI::datashield.assign.resource(conns[4], "resource_expr",
"OMICS.genexpr_MOB")
DSI::datashield.assign.resource(conns[5], "resource_expr",
"OMICS.genexpr_RHE")
DSI::datashield.assign.resource(conns[6], "resource_expr",
"OMICS.genexpr_SAB")With these instructions, we have assigned the resources into our remote R session to a variable called “resource_expr”. To verify this step has been performed correctly, we could use the ds.class() function to check for their class and that they exist on the remote session. Then, we have to resolve the resource and retrieve the data in the remote session. For that, we will use the DSI::datashield.assign.expr() function.
# We resolve the resource
DSI::datashield.assign.expr(conns = conns, symbol = "resource_expr_HELIX",
expr = as.symbol("as.resource.object(resource_expr)"))As a result, we will get an R object (here named “resource_expr_HELIX”) containing the available “ExpressionSets” files for cohorts. For created objects, we can extract some useful information including the class, and the name of the genes included in the microarray and additional phenotype data available.
# Retrieve the class of loaded files:
ds.class("resource_expr_HELIX")
# Get the names of the first features (probes) available for the first cohort:
name.features <- ds.featureNames("resource_expr_HELIX")
lapply(name.features, head)[[1]]
# Get the names of the phenotype data available for the first cohort:
name.phenotypes <- ds.varLabels("resource_expr_HELIX")
lapply(name.phenotypes, head)[[1]]
# Also the annotation information can be inspected by:
name.annot <- ds.fvarLabels("resource_expr_HELIX")
lapply(name.annot, head)[[1]]
# Annotation information refers to additional data available at the feature
# level and that could be added to the summary statistics for a better
# identification of significant genes.11.3 Differential gene expression analysis.
The differential expression analysis in dsOmicsClient is implemented in the funcion ds.limma(). Although, by default, the function considers that data is obtained from a microarray experiment (type.data = “microarray”), it could also be applied to RNAseq data.
In this case, since we are interested in evaluating the gene expression differences between boys and girls, we will run the next model (adjusted by possible confounders such as age and ethnicity):
model.sex <- ds.limma(model = ~ e3_sex + age_sample_years + h_ethnicity_cauc,
Set = "resource_expr_HELIX")The top differentially expressed genes by cohort can then be visualized by:
model.sex$BIB
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC13001096.hg.1 162 -0.126 0.164 -7.09 4.12e-11 0.00000119
2 TC22000217.hg.1 162 -0.156 0.0180 -6.74 2.72e-10 0.00000391
3 TC17000757.hg.1 162 0.0900 0.0233 6.22 4.23e- 9 0.0000387
4 TC20000999.hg.1 162 0.172 0.0242 6.10 7.48e- 9 0.0000387
5 TC20001182.hg.1 162 0.411 0.0267 6.10 7.58e- 9 0.0000387
6 TC20000178.hg.1 162 0.341 0.0321 6.09 8.09e- 9 0.0000387
7 TC19001398.hg.1 162 -0.207 0.0258 -5.79 3.68e- 8 0.000151
8 TC04001402.hg.1 162 -0.296 0.0459 -5.75 4.30e- 8 0.000154
9 TC05001973.hg.1 162 -0.390 0.0231 -5.52 1.34e- 7 0.000429
10 TC09000119.hg.1 162 0.0775 0.0309 5.49 1.57e- 7 0.000452
# ... with 28,728 more rows
$EDEN
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC12000357.hg.1 109 -0.0574 0.127 -4.71 0.00000729 0.121
2 TC15000639.hg.1 109 0.135 0.0200 4.64 0.00000985 0.121
3 TC02004993.hg.1 109 0.124 0.0302 4.52 0.0000158 0.121
4 TC02001326.hg.1 109 -0.0736 0.0418 -4.51 0.0000168 0.121
5 TC15001330.hg.1 109 0.0945 0.0488 4.34 0.0000317 0.135
6 TC19001398.hg.1 109 -0.191 0.0335 -4.27 0.0000416 0.135
7 TC20000999.hg.1 109 0.115 0.0279 4.26 0.0000441 0.135
8 TC12002776.hg.1 109 -0.155 0.0504 -4.25 0.0000459 0.135
9 TC05000721.hg.1 109 -0.171 0.0266 -4.23 0.0000491 0.135
10 TC01006291.hg.1 109 0.113 0.0315 4.23 0.0000498 0.135
# ... with 28,728 more rows
$KANC
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC11001104.hg.1 151 -0.346 0.148 -6.57 7.84e-10 0.0000225
2 TC10000002.hg.1 151 0.0839 0.0192 6.34 2.51e- 9 0.0000360
3 TC01001596.hg.1 151 -0.224 0.0245 -5.93 1.96e- 8 0.000158
4 TC04001402.hg.1 151 -0.226 0.0272 -5.91 2.20e- 8 0.000158
5 TC01000909.hg.1 151 -0.147 0.0510 -5.76 4.53e- 8 0.000248
6 TC15000872.hg.1 151 0.102 0.0409 5.72 5.44e- 8 0.000248
7 TC08000814.hg.1 151 0.173 0.0296 5.70 6.03e- 8 0.000248
8 TC13000150.hg.1 151 -0.0825 0.0424 -5.66 7.56e- 8 0.000272
9 TC20000178.hg.1 151 0.257 0.0298 5.55 1.27e- 7 0.000394
10 TC05001973.hg.1 151 -0.294 0.0292 -5.53 1.37e- 7 0.000394
# ... with 28,728 more rows
$MoBA
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC19001274.hg.1 245 -0.291 0.136 -7.19 8.03e-12 0.000000231
2 TC13000150.hg.1 245 -0.0768 0.0125 -6.99 2.68e-11 0.000000385
3 TC06000733.hg.1 245 0.110 0.0184 6.81 7.63e-11 0.000000563
4 TC15001330.hg.1 245 0.0911 0.0194 6.80 7.84e-11 0.000000563
5 TC01001720.hg.1 245 -0.262 0.0229 -6.75 1.07e-10 0.000000617
6 TC19001398.hg.1 245 -0.229 0.0277 -6.56 3.19e-10 0.00000142
7 TC01001596.hg.1 245 -0.204 0.0187 -6.55 3.46e-10 0.00000142
8 TC09000119.hg.1 245 0.0859 0.0368 6.47 5.21e-10 0.00000187
9 TC10002872.hg.1 245 0.0882 0.0192 6.39 8.39e-10 0.00000268
10 TC10000002.hg.1 245 0.0677 0.0243 6.27 1.62e- 9 0.00000466
# ... with 28,728 more rows
$Rhea
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC09001482.hg.1 156 0.108 0.150 7.17 2.77e-11 0.000000487
2 TC10000002.hg.1 156 0.0843 0.0201 7.14 3.39e-11 0.000000487
3 TC12003242.hg.1 156 0.132 0.0220 6.52 9.25e-10 0.00000886
4 TC15000872.hg.1 156 0.128 0.0209 6.12 7.39e- 9 0.0000531
5 TC17000757.hg.1 156 0.0990 0.0279 5.77 4.24e- 8 0.000244
6 TC11002451.hg.1 156 0.0894 0.0380 5.63 8.26e- 8 0.000367
7 TC20001182.hg.1 156 0.351 0.0225 5.61 8.94e- 8 0.000367
8 TC20000501.hg.1 156 0.0949 0.0540 5.36 2.93e- 7 0.00105
9 TC01000598.hg.1 156 0.0911 0.0234 5.29 4.03e- 7 0.00129
10 TC13000150.hg.1 156 -0.0741 0.0375 -5.22 5.54e- 7 0.00159
# ... with 28,728 more rows
$INMASAB
# A tibble: 28,738 x 7
id n beta SE t P.Value adj.P.Val
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC03003362.hg.1 184 -0.0779 0.127 -6.57 5.11e-10 0.0000147
2 TC09000450.hg.1 184 0.0782 0.0151 6.35 1.61e- 9 0.0000193
3 TC01006291.hg.1 184 0.114 0.0199 6.31 2.02e- 9 0.0000193
4 TC03000563.hg.1 184 -0.200 0.0225 -6.18 3.96e- 9 0.0000284
5 TC19001398.hg.1 184 -0.219 0.0319 -6.07 7.08e- 9 0.0000407
6 TC10002953.hg.1 184 0.0983 0.0333 5.62 6.96e- 8 0.000322
7 TC01000909.hg.1 184 -0.129 0.0247 -5.60 7.84e- 8 0.000322
8 TC17000757.hg.1 184 0.0793 0.0417 5.40 2.00e- 7 0.000709
9 TC10002872.hg.1 184 0.0917 0.0225 5.38 2.22e- 7 0.000709
10 TC01003403.hg.1 184 -0.182 0.0331 -5.32 2.97e- 7 0.000855
# ... with 28,728 more rows
attr(,"class")
[1] "dsLimma" "list" We can verify whether the distribution of the observed p-values are the ones we would expect in this type of analyses:
ggplot(model.sex$BIB, aes(x=P.Value))+
geom_histogram(color="darkblue", fill="lightblue")+labs(
title="P-Value histogram plot",x="Raw p-value gender effect", y = "Count")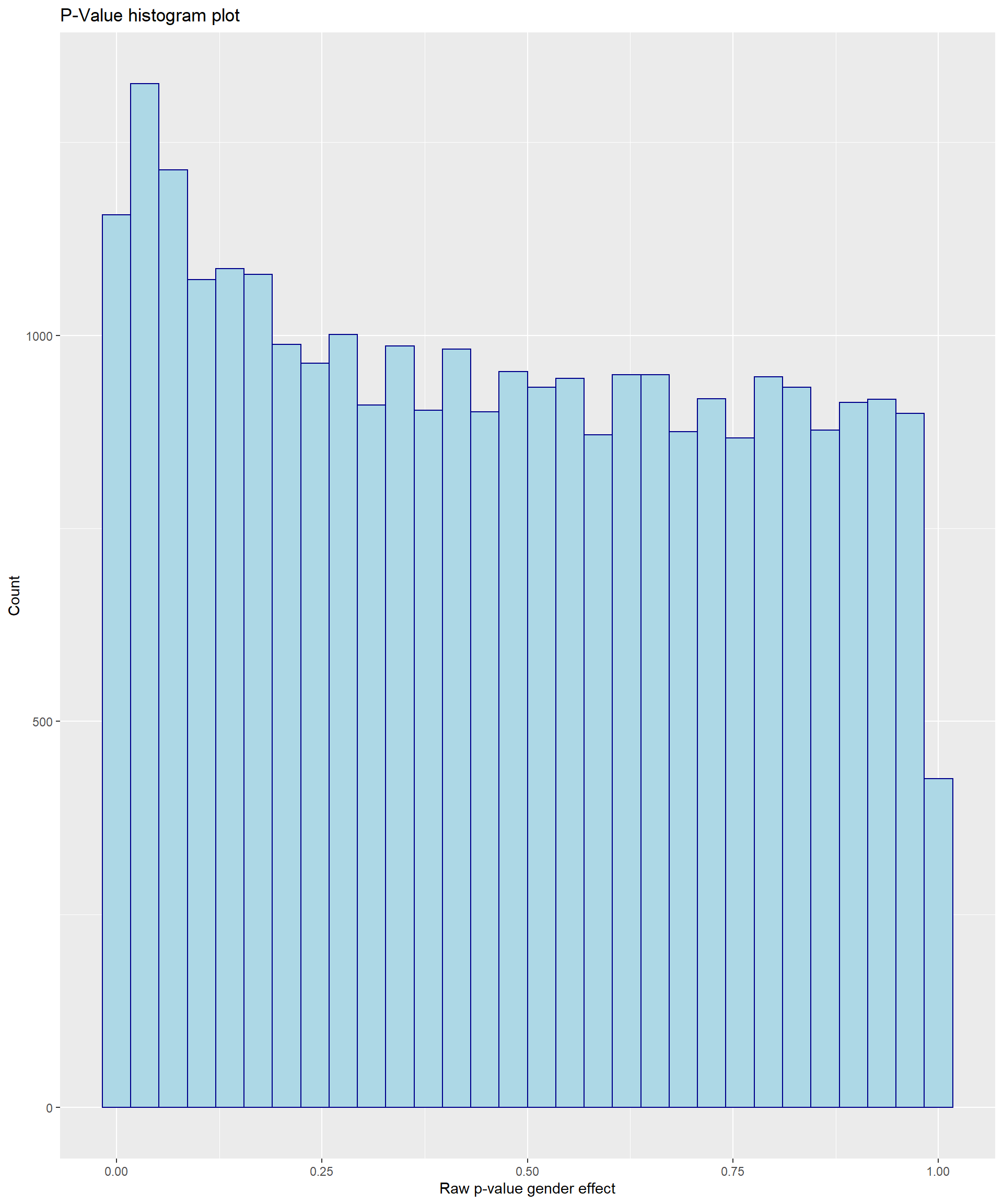 Another functionality of dsOmicsClient is that we can add annotation information at the probe-level to the output that is obtained with ds.limma(). Annotation information will be available as part of the expression set file “resource_expr_HELIX” and can be inspected by:
Another functionality of dsOmicsClient is that we can add annotation information at the probe-level to the output that is obtained with ds.limma(). Annotation information will be available as part of the expression set file “resource_expr_HELIX” and can be inspected by:
# Get annotation information for the first cohort
ds.fvarLabels("resource_expr_HELIX")[[1]] [1] "transcript_cluster_id" "probeset_id" "seqname"
[4] "strand" "start" "stop"
[7] "total_probes" "gene_assignment" "mrna_assignment"
[10] "notes" "phase" "TC_size"
[13] "TSS_Affy" "EntrezeGeneID_Affy" "GeneSymbol_Affy"
[16] "GeneSymbolDB" "GeneSymbolDB2" "mrna_ID"
[19] "mrna_DB" "mrna_N" "notesYN"
[22] "geneYN" "genes_N" "CallRate"
[25] "fil1" In case we were interested in adding gene symbol, entrezID and chromosome number mapping probes from each output, we can run:
model.sex_annot <- ds.limma(model = ~ e3_sex + age_sample_years + h_ethnicity_cauc,
Set = "resource_expr_HELIX",
annotCols = c("GeneSymbol_Affy","seqname",
"EntrezeGeneID_Affy"))The results for each of the cohort (ordered by level of significance) can be accessed by:
model.sex_annot$BIB
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC1300~ 162 -0.126 0.164 -7.09 4.12e-11 1.19e-6 INTS6-AS1 chr13
2 TC2200~ 162 -0.156 0.0180 -6.74 2.72e-10 3.91e-6 OSBP2 chr22
3 TC1700~ 162 0.0900 0.0233 6.22 4.23e- 9 3.87e-5 TLK2 chr17
4 TC2000~ 162 0.172 0.0242 6.10 7.48e- 9 3.87e-5 SYCP2 chr20
5 TC2000~ 162 0.411 0.0267 6.10 7.58e- 9 3.87e-5 FRG1BP chr20
6 TC2000~ 162 0.341 0.0321 6.09 8.09e- 9 3.87e-5 FRG1BP;FRG1CP chr20
7 TC1900~ 162 -0.207 0.0258 -5.79 3.68e- 8 1.51e-4 RPS4XP21 chr19
8 TC0400~ 162 -0.296 0.0459 -5.75 4.30e- 8 1.54e-4 TSPAN5 chr4
9 TC0500~ 162 -0.390 0.0231 -5.52 1.34e- 7 4.29e-4 FAXDC2 chr5
10 TC0900~ 162 0.0775 0.0309 5.49 1.57e- 7 4.52e-4 IFT74 chr9
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
$EDEN
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC12000~ 109 -0.0574 0.127 -4.71 7.29e-6 0.121 CACNB3 chr12
2 TC15000~ 109 0.135 0.0200 4.64 9.85e-6 0.121 RPLP1 chr15
3 TC02004~ 109 0.124 0.0302 4.52 1.58e-5 0.121 C2orf15 chr2
4 TC02001~ 109 -0.0736 0.0418 -4.51 1.68e-5 0.121 DES chr2
5 TC15001~ 109 0.0945 0.0488 4.34 3.17e-5 0.135 MYEF2 chr15
6 TC19001~ 109 -0.191 0.0335 -4.27 4.16e-5 0.135 RPS4XP21 chr19
7 TC20000~ 109 0.115 0.0279 4.26 4.41e-5 0.135 SYCP2 chr20
8 TC12002~ 109 -0.155 0.0504 -4.25 4.59e-5 0.135 ART4 chr12
9 TC05000~ 109 -0.171 0.0266 -4.23 4.91e-5 0.135 IGIP chr5
10 TC01006~ 109 0.113 0.0315 4.23 4.98e-5 0.135 CFH chr1
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
$KANC
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC1100~ 151 -0.346 0.148 -6.57 7.84e-10 0.0000225 TBCEL chr11
2 TC1000~ 151 0.0839 0.0192 6.34 2.51e- 9 0.0000360 ZMYND11 chr10
3 TC0100~ 151 -0.224 0.0245 -5.93 1.96e- 8 0.000158 SWT1 chr1
4 TC0400~ 151 -0.226 0.0272 -5.91 2.20e- 8 0.000158 TSPAN5 chr4
5 TC0100~ 151 -0.147 0.0510 -5.76 4.53e- 8 0.000248 S1PR1 chr1
6 TC1500~ 151 0.102 0.0409 5.72 5.44e- 8 0.000248 MAN2A2 chr15
7 TC0800~ 151 0.173 0.0296 5.70 6.03e- 8 0.000248 LY6E chr8
8 TC1300~ 151 -0.0825 0.0424 -5.66 7.56e- 8 0.000272 DGKH chr13
9 TC2000~ 151 0.257 0.0298 5.55 1.27e- 7 0.000394 FRG1BP;FRG1CP chr20
10 TC0500~ 151 -0.294 0.0292 -5.53 1.37e- 7 0.000394 FAXDC2 chr5
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
$MoBA
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC1900~ 245 -0.291 0.136 -7.19 8.03e-12 2.31e-7 PLVAP chr19
2 TC1300~ 245 -0.0768 0.0125 -6.99 2.68e-11 3.85e-7 DGKH chr13
3 TC0600~ 245 0.110 0.0184 6.81 7.63e-11 5.63e-7 DDX43 chr6
4 TC1500~ 245 0.0911 0.0194 6.80 7.84e-11 5.63e-7 MYEF2 chr15
5 TC0100~ 245 -0.262 0.0229 -6.75 1.07e-10 6.17e-7 CTSE chr1
6 TC1900~ 245 -0.229 0.0277 -6.56 3.19e-10 1.42e-6 RPS4XP21 chr19
7 TC0100~ 245 -0.204 0.0187 -6.55 3.46e-10 1.42e-6 SWT1 chr1
8 TC0900~ 245 0.0859 0.0368 6.47 5.21e-10 1.87e-6 IFT74 chr9
9 TC1000~ 245 0.0882 0.0192 6.39 8.39e-10 2.68e-6 FAM24B;CUZD1 chr10
10 TC1000~ 245 0.0677 0.0243 6.27 1.62e- 9 4.66e-6 ZMYND11 chr10
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
$Rhea
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC0900~ 156 0.108 0.150 7.17 2.77e-11 4.87e-7 KIAA0368 chr9
2 TC1000~ 156 0.0843 0.0201 7.14 3.39e-11 4.87e-7 ZMYND11 chr10
3 TC1200~ 156 0.132 0.0220 6.52 9.25e-10 8.86e-6 PCBP2 chr12
4 TC1500~ 156 0.128 0.0209 6.12 7.39e- 9 5.31e-5 MAN2A2 chr15
5 TC1700~ 156 0.0990 0.0279 5.77 4.24e- 8 2.44e-4 TLK2 chr17
6 TC1100~ 156 0.0894 0.0380 5.63 8.26e- 8 3.67e-4 ZBTB44 chr11
7 TC2000~ 156 0.351 0.0225 5.61 8.94e- 8 3.67e-4 FRG1BP chr20
8 TC2000~ 156 0.0949 0.0540 5.36 2.93e- 7 1.05e-3 GID8 chr20
9 TC0100~ 156 0.0911 0.0234 5.29 4.03e- 7 1.29e-3 CMPK1 chr1
10 TC1300~ 156 -0.0741 0.0375 -5.22 5.54e- 7 1.59e-3 DGKH chr13
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
$INMASAB
# A tibble: 28,738 x 10
id n beta SE t P.Value adj.P.Val GeneSymbol_Affy seqname
<chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 TC0300~ 184 -0.0779 0.127 -6.57 5.11e-10 0.0000147 PHLDB2 chr3
2 TC0900~ 184 0.0782 0.0151 6.35 1.61e- 9 0.0000193 PHF2 chr9
3 TC0100~ 184 0.114 0.0199 6.31 2.02e- 9 0.0000193 CFH chr1
4 TC0300~ 184 -0.200 0.0225 -6.18 3.96e- 9 0.0000284 CD96 chr3
5 TC1900~ 184 -0.219 0.0319 -6.07 7.08e- 9 0.0000407 RPS4XP21 chr19
6 TC1000~ 184 0.0983 0.0333 5.62 6.96e- 8 0.000322 CUZD1 chr10
7 TC0100~ 184 -0.129 0.0247 -5.60 7.84e- 8 0.000322 S1PR1 chr1
8 TC1700~ 184 0.0793 0.0417 5.40 2.00e- 7 0.000709 TLK2 chr17
9 TC1000~ 184 0.0917 0.0225 5.38 2.22e- 7 0.000709 FAM24B;CUZD1 chr10
10 TC0100~ 184 -0.182 0.0331 -5.32 2.97e- 7 0.000855 SLAMF6 chr1
# ... with 28,728 more rows, and 1 more variable: EntrezeGeneID_Affy <chr>
attr(,"class")
[1] "dsLimma" "list" 11.4 Surrogate variable analysis.
We can also check whether there is inflation just executing:
qqplot(model.sex$BIB$P.Value)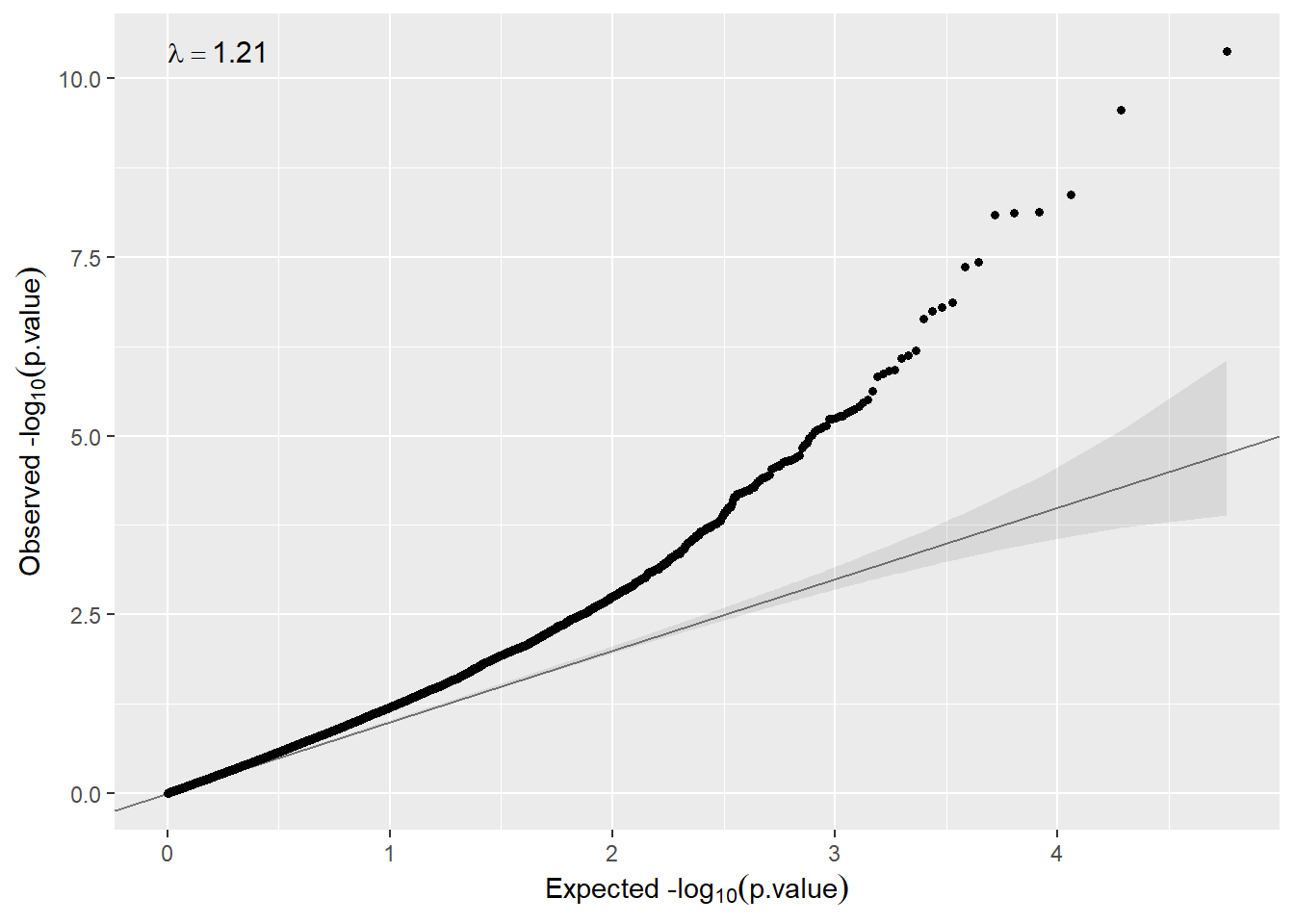
So, in that case, the model does not need to remove unwanted variability (λ<2). If that was not the case, we could use surrogate variable analysis just changing the argument sva=TRUE..
The ds.limma function has another arguments that can be used to fit other type of models:
- method: Linear regression (“ls”) or robust regression (“robust”) used in limma (lmFit).
- robust: Logical. Robust method used for outlier sample variances used in limma (eBayes).
- normalization: normalization method used in the voom transformation (default “none”).
- voomQualityWeights: Logical. Should voomQualityWeights function be used instead of voom? (default FALSE)
- big: Logical. If sva was set to TRUE, should SmartSVA be implemented?
Then, data can be combined meta-analysed as follows:
metaP.model.sex.annot <- metaPvalues(model.sex_annot)
metaP.model.sex.annot# A tibble: 28,738 x 8
id BIB EDEN KANC MoBA Rhea INMASAB p.meta
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 TC1000000~ 3.10e- 6 8.20e-4 2.51e-9 1.62e- 9 3.39e-11 1.09e-5 2.63e-34
2 TC1900139~ 3.68e- 8 4.16e-5 1.92e-7 3.19e-10 6.26e- 5 7.08e-9 2.54e-33
3 TC2000118~ 7.58e- 9 7.07e-5 5.48e-7 8.83e- 9 8.94e- 8 8.94e-7 1.16e-32
4 TC1700075~ 4.23e- 9 7.24e-4 9.59e-6 3.60e- 9 4.24e- 8 2.00e-7 4.64e-32
5 TC1500087~ 4.20e- 6 6.67e-5 5.44e-8 3.83e- 8 7.39e- 9 8.93e-7 1.84e-31
6 TC2000017~ 8.09e- 9 9.86e-5 1.27e-7 9.23e- 8 1.26e- 6 1.46e-6 7.53e-31
7 TC1300109~ 4.12e-11 1.01e-4 3.58e-5 3.58e- 7 1.02e- 6 1.88e-5 3.56e-29
8 TC0100090~ 5.82e- 5 8.74e-4 4.53e-8 3.43e- 9 2.77e- 6 7.84e-8 5.75e-29
9 TC0900148~ 7.50e- 6 1.11e-3 1.74e-5 2.15e- 6 2.77e-11 8.15e-7 2.17e-28
10 TC1300015~ 5.68e- 6 8.56e-3 7.56e-8 2.68e-11 5.54e- 7 1.97e-4 3.22e-28
# ... with 28,728 more rows# Get the number of differentially expressed genes in the meta approach
dim(metaP.model.sex.annot[metaP.model.sex.annot$p.meta<0.05,])[1] 3839 811.5 Enrichment analysis of functional annotations.
Once we have obtained the top differentially expressed genes per cohort, we could extract their gene symbols or gene entrez ids directly from each output dataframe and continue with the functional annotation analysis in our local session as usual. To this end, one of the classic approaches is to evaluate whether any information (in the form of annotations) is over-represented in the gene list compared to the rest of the genes in the genome. This type of enrichment analysis is based on annotating genes with information available in different databases such as Gene Ontology or the Kyoto Encyclopedia of Genes and Genomes, and establishing the frequencies of each term in the gene list and the rest of the genome. This allows a statistical test to be applied to determine which functional annotations are significantly enriched in the list.
To extract the list of top differentially expressed genes by cohort and perform enrichment analysis with KEGG we can run the next code lines:
# Get the entrez id for the top differentially expressed genes in each cohort.
get_entrezid <- function(x){ x[(x$adj.P.Val < 0.05),c("id","EntrezeGeneID_Affy")] }
topgenes_by_cohort <- lapply(model.sex_annot,get_entrezid)
length(unlist(lapply(topgenes_by_cohort,function(x) x[,2])))[1] 666# Get the entrez id for the top differentially expressed genes in the meta-analysis
# approach
ensembl <- useMart("ensembl", dataset = "hsapiens_gene_ensembl")
filters <- listFilters(ensembl)
attributes <- listAttributes(ensembl)
affy_id <- gsub(x = as.character(metaP.model.sex.annot[metaP.model.sex.annot$p.meta<
0.05/nrow(metaP.model.sex.annot),1]$id), pattern = "hg.1", "hg")
# Number of probes passing multiple-testing correction in the meta-analysis.
length(affy_id)[1] 325# Get the entrez id of the genes mapped by the probes from the previous list.
topgenes_metaP <- getBM(attributes = c("entrezgene_id"),
filters = "affy_hta_2_0",
values = affy_id,
mart = ensembl)
length(topgenes_metaP[,1])[1] 276# Functional enrichment analysis with KEGG.
# We will use as reference a list of genes loaded from the DOSE package.
data(geneList, package="DOSE")
# Enrichment for top genes from each cohort
enrichKEGG <- enrichKEGG(gene = unlist(lapply(topgenes_by_cohort,function(x) x[,2])),
organism = 'hsa',
pvalueCutoff = 0.1)
head(enrichKEGG) ID Description GeneRatio
hsa03010 hsa03010 Ribosome 20/183
hsa03008 hsa03008 Ribosome biogenesis in eukaryotes 16/183
hsa04211 hsa04211 Longevity regulating pathway 10/183
hsa04213 hsa04213 Longevity regulating pathway - multiple species 7/183
hsa04150 hsa04150 mTOR signaling pathway 11/183
hsa04152 hsa04152 AMPK signaling pathway 9/183
BgRatio pvalue p.adjust qvalue
hsa03010 158/8135 2.903164e-10 7.490164e-08 6.295282e-08
hsa03008 109/8135 2.144118e-09 2.765912e-07 2.324675e-07
hsa04211 89/8135 2.831858e-05 2.435398e-03 2.046887e-03
hsa04213 62/8135 4.497217e-04 2.900705e-02 2.437965e-02
hsa04150 156/8135 7.577933e-04 3.910213e-02 3.286430e-02
hsa04152 120/8135 1.483310e-03 5.812931e-02 4.885613e-02
geneID
hsa03010 6176/6171/6208/100169751/100169753/100169754/100169755/100169756/100169757/100169758/100169759/100169761/100169762/100169763/100169764/100169765/100169766/100169767/100169768/23521
hsa03008 100169751/100169753/100169754/100169755/100169756/100169757/100169758/100169759/100169761/100169762/100169763/100169764/100169765/100169766/100169767/100169768
hsa04211 5567/5562/6198/27244/9586/64764/6648/3480/10000/207
hsa04213 5567/5562/6198/6648/3480/10000/207
hsa04150 523/6446/5562/6198/7477/1975/3480/5578/10000/207/9706
hsa04152 5562/6198/55844/1938/9586/64764/3480/10000/207
Count
hsa03010 20
hsa03008 16
hsa04211 10
hsa04213 7
hsa04150 11
hsa04152 9# Enrichment for top genes from the meta-analysis
enrichKEGG_meta <- enrichKEGG(gene = as.character(topgenes_metaP[,1]),
organism = 'hsa',
pvalueCutoff = 0.1)
head(enrichKEGG_meta) ID Description GeneRatio BgRatio pvalue
hsa04211 hsa04211 Longevity regulating pathway 7/117 89/8135 0.0002767746
p.adjust qvalue geneID Count
hsa04211 0.06393493 0.0614731 9586/6198/3480/26060/27244/5562/5567 7# The pathways can be visualized alongside the genes associated with them:
browseKEGG(enrichKEGG, 'hsa04211')As a result, we identify a list of 325 probes (mapping 287 genes) differentially expressed between boys and girls of the HELIX cohort, passing multiple-testing correction filter (P-value threshold=1.74e-06). As a proof of the ability of OmicSHIELD to be integrated with other R functionalities and bioconductor utilties, we continued the showcase presenting a functional enrichment analysis (FEA) of results, showing that significant DE genes participate in processes with evident and previously-described sexual dimorphism such is the case of “Longevity regulating pathways” (hsa04211).
Once analyses are completed, we proceed to close the DataSHIELD session by:
datashield.logout(conns)