7 PLINK
A simple association test use case will be illustrated in this section to portrait the usage of PLINK in OmicSHIELD.
|
⚠️ RESOURCES USED ALONG THIS SECTION |
||||||
|---|---|---|---|---|---|---|
|
From https://opal-demo.obiba.org/ : |
||||||
|
The structure followed is illustrated on the following figure.
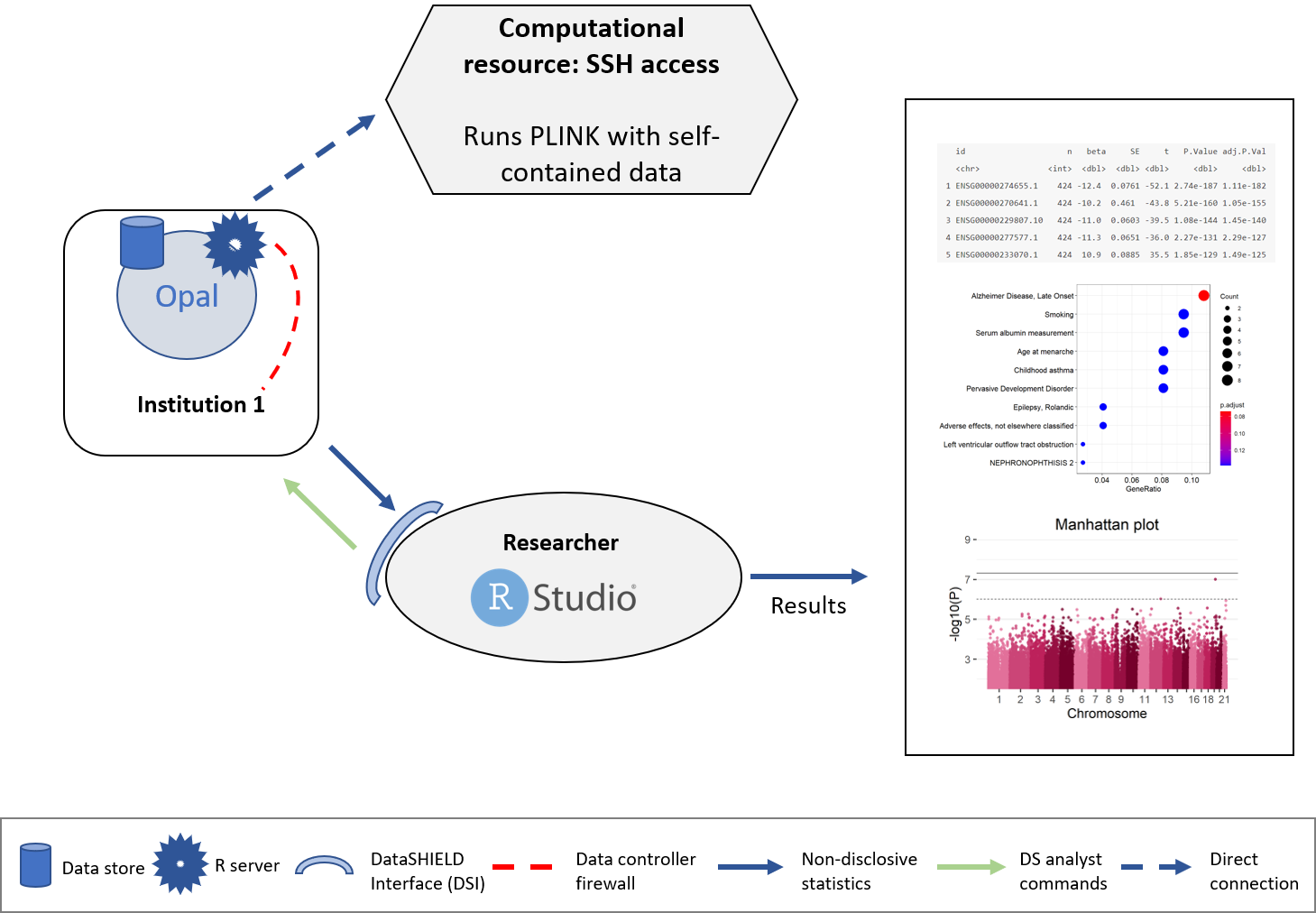
Figure 7.1: Proposed infrastructure for PLINK analysis.
The data analyst corresponds to the “RStudio” session, which through DataSHIELD Interface (DSI) connects with the Opal server located at the cohort network. This Opal server contains a resource with the URL where the computational resource is located, also the access credentials are kept secret inside the resource and only the Opal administrator has access to them. A connection to this computational resource is created on the Opal server and the aggregated results are passed to the analyst.
7.1 Connection to the Opal server
We have to create an Opal connection object to the cohort server. We do that using the following functions.
library(DSOpal)
library(dsBaseClient)
library(dsOmicsClient)
builder <- newDSLoginBuilder()
builder$append(server = "study1", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "P@ssw0rd",
resource = "RSRC.brge_plink",
profile = "omics")
logindata <- builder$build()
conns <- datashield.login(logins = logindata, assign = TRUE,
symbol = "client")7.2 Build the PLINK call
Now, we are ready to run any PLINK command from the client site. Notice that in this case we want to assess association between the genotype data in bed format and use as phenotype the variable ‘asthma’ that is in the file ‘brge.phe’ in the 6th column. The sentence in a PLINK command would be (NOTE: we avoid –out to indicate the output file since the file will be available in R as a tibble). The arguments must be encapsulated in a single character without the command ‘plink’.
plink.arguments <- "--bfile brge --logistic --pheno brge.phe --mpheno 6 --covar brge.phe --covar-name gender,age"7.3 Call the analysis
Then, the analyses are performed by:
ans.plink <- ds.PLINK("client", plink.arguments)The object ans.plink contains the PLINK results at each server as well as the outuput provided by PLINK
lapply(ans.plink, names)$study1
[1] "results" "plink.out" head(ans.plink$study1$results)# A tibble: 6 x 10
CHR SNP BP A1 TEST NMISS OR STAT P X10
<dbl> <chr> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
1 0 MitoC3993T 3993 T ADD 2286 0.752 -1.33 0.182 <NA>
2 0 MitoC3993T 3993 T gender 2286 0.742 -3.27 0.00107 <NA>
3 0 MitoC3993T 3993 T age 2286 1.00 0.565 0.572 <NA>
4 0 MitoG4821A 4821 A ADD 2282 2.68 1.71 0.0879 <NA>
5 0 MitoG4821A 4821 A gender 2282 0.740 -3.31 0.000940 <NA>
6 0 MitoG4821A 4821 A age 2282 1.00 0.465 0.642 <NA> ans.plink$study$plink.out$status
[1] 0
$output
[1] ""
[2] "@----------------------------------------------------------@"
[3] "| PLINK! | v1.07 | 10/Aug/2009 |"
[4] "|----------------------------------------------------------|"
[5] "| (C) 2009 Shaun Purcell, GNU General Public License, v2 |"
[6] "|----------------------------------------------------------|"
[7] "| For documentation, citation & bug-report instructions: |"
[8] "| http://pngu.mgh.harvard.edu/purcell/plink/ |"
[9] "@----------------------------------------------------------@"
[10] ""
[11] "Skipping web check... [ --noweb ] "
[12] "Writing this text to log file [ /tmp/ssh-7484/out.log ]"
[13] "Analysis started: Mon Apr 4 15:55:21 2022"
[14] ""
[15] "Options in effect:"
[16] "\t--bfile brge"
[17] "\t--logistic"
[18] "\t--pheno brge.phe"
[19] "\t--mpheno 6"
[20] "\t--covar brge.phe"
[21] "\t--covar-name gender,age"
[22] "\t--noweb"
[23] "\t--out /tmp/ssh-7484/out"
[24] ""
[25] "Reading map (extended format) from [ brge.bim ] "
[26] "100000 markers to be included from [ brge.bim ]"
[27] "Reading pedigree information from [ brge.fam ] "
[28] "2312 individuals read from [ brge.fam ] "
[29] "2312 individuals with nonmissing phenotypes"
[30] "Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)"
[31] "Missing phenotype value is also -9"
[32] "725 cases, 1587 controls and 0 missing"
[33] "1097 males, 1215 females, and 0 of unspecified sex"
[34] "Reading genotype bitfile from [ brge.bed ] "
[35] "Detected that binary PED file is v1.00 SNP-major mode"
[36] "Reading alternate phenotype from [ brge.phe ] "
[37] "2312 individuals with non-missing alternate phenotype"
[38] "Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)"
[39] "Missing phenotype value is also -9"
[40] "725 cases, 1587 controls and 0 missing"
[41] "Reading 6 covariates from [ brge.phe ] with nonmissing values for 2199 individuals"
[42] "Selected subset of 2 from 6 covariates"
[43] "For these, nonmissing covariate values for 2312 individuals"
[44] "Before frequency and genotyping pruning, there are 100000 SNPs"
[45] "2312 founders and 0 non-founders found"
[46] "6009 heterozygous haploid genotypes; set to missing"
[47] "Writing list of heterozygous haploid genotypes to [ /tmp/ssh-7484/out.hh ]"
[48] "7 SNPs with no founder genotypes observed"
[49] "Warning, MAF set to 0 for these SNPs (see --nonfounders)"
[50] "Writing list of these SNPs to [ /tmp/ssh-7484/out.nof ]"
[51] "Total genotyping rate in remaining individuals is 0.994408"
[52] "0 SNPs failed missingness test ( GENO > 1 )"
[53] "0 SNPs failed frequency test ( MAF < 0 )"
[54] "After frequency and genotyping pruning, there are 100000 SNPs"
[55] "After filtering, 725 cases, 1587 controls and 0 missing"
[56] "After filtering, 1097 males, 1215 females, and 0 of unspecified sex"
[57] "Converting data to Individual-major format"
[58] "Writing logistic model association results to [ /tmp/ssh-7484/out.assoc.logistic ] "
[59] ""
[60] "Analysis finished: Mon Apr 4 15:57:15 2022"
[61] ""
$error
character(0)
$command
[1] "cd /home/master/brge && plink1 --bfile brge --logistic --pheno brge.phe --mpheno 6 --covar brge.phe --covar-name gender,age --noweb --out /tmp/ssh-7484/out"
attr(,"class")
[1] "resource.exec"datashield.logout(conns)