11 Setup
As described in a previous Chapter the resourcer R package allows users to deal with the main data sources (using tidyverse, DBI, dplyr, sparklyr, MongoDB, AWS S3, SSH etc.) and is easily extensible to new ones including specific data infrastructure in R or Bioconductor. So far ExpressionSet and RangedSummarizedExperiment objects saved in .RData files are accesible through the resourcer package. The dsOmics package contains a new extension that deals with VCF (Variant Calling Format) files which are coerced to a GDS (Genomic Data Storage) format (VCF2GDS).
In order to achieve this resourcer extension, two R6 classes have been implemented:
GDSFileResourceResolverclass which handles file-base resources with data in GDS or VCF formats. This class is responsible for creating aGDSFileResourceClientobject instance from an assigned resource.GDSFileResourceClientclass which is responsible for getting the referenced file and making a connection (created byGWASTools) to the GDS file (will also convert the VCF file to a GDS file on the fly, usingSNPRelate). For the subsequent analysis, it is this connection handle to the GDS file that will be used.
11.1 Providing DataSHIELD packages in the opal server
The required DataSHIELD packages must be uploaded to Opal through the Administration site by accessing to DataSHIELD tab. In our case, both dsBase and dsOmics and resourcer packages must be installed as is illustrated in the figure.
Figure 11.1: Installed packages in the demo opal server
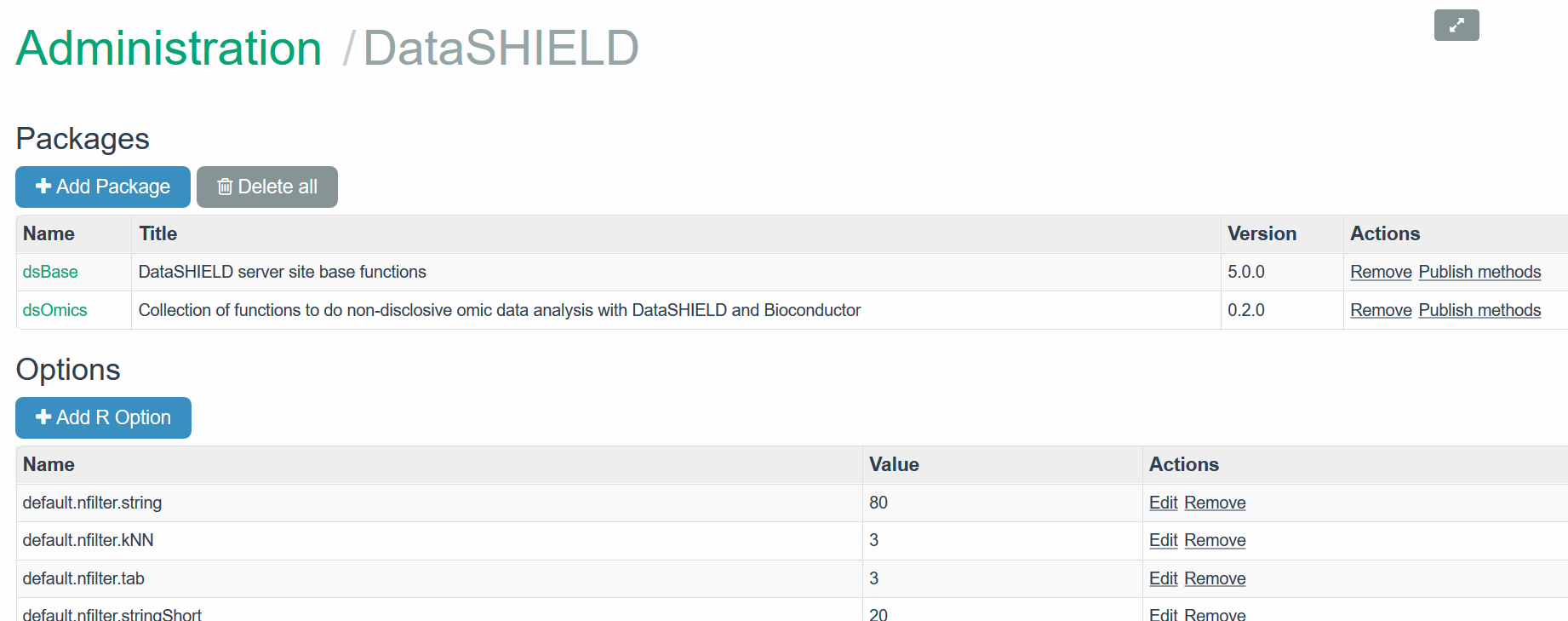
The tab +Add package can be used to install a new package. The figure depicts how dsOmics was intalled on Opal
Figure 11.2: Description how dsOmics package was intalled into the demo opal server
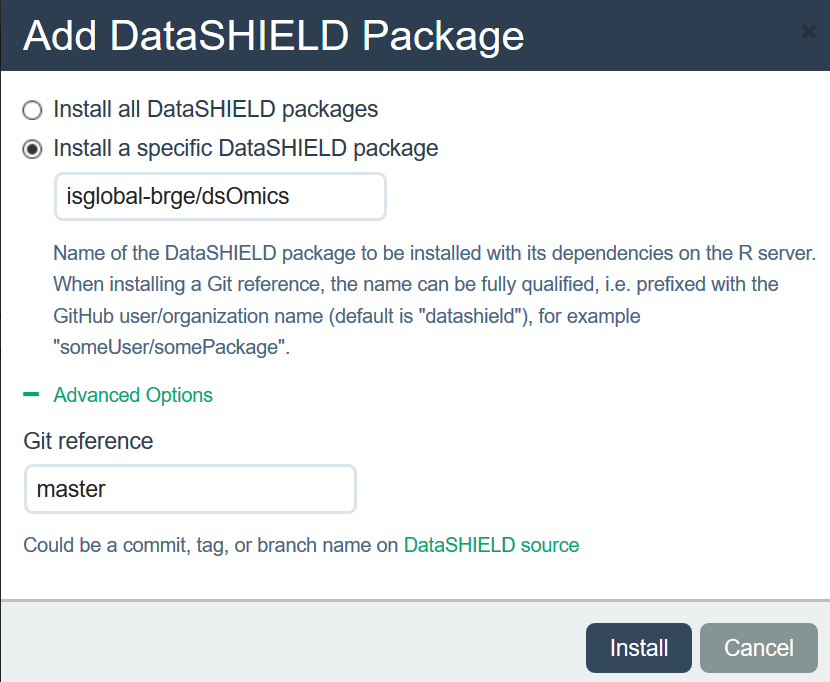
For reproducing this book the following packages must be installed on Opal
From CRAN:
- resourcer
From Github:
- datashield/dsBase
- datashield/dsGeo (tombisho/dsGeo)
- isglobal-brge/dsOmicsNote that the dsGeo package imports the sp, rgeos and rgdal R packages. rgeos and rgdal in turn require some additional libraries which can be installed as follows (on Ubuntu systems - see the notes in rgeos and rgdal for other operating systems):
sudo apt-get update
sudo apt-get install libgdal-dev libproj-dev libgeos++dev11.2 Required R Packages in the client site (e.g. local machine)
Using DataSHIELD also requires some R packages to be installed on the client site. So far, the following R packages must be installed (in their development version):
install.packages("DSOpal", dependencies = TRUE)
install.packages("dsBaseClient", repos = c("https://cloud.r-project.org", "https://cran.obiba.org"), dependencies = TRUE)
devtools::install_github("isglobal-brge/dsOmicsClient", dependencies = TRUE)
devtools::install_github("tombisho/dsGeoClient", dependencies = TRUE)The package dependencies are then loaded as follows: