7 The resourcer Package
7.1 Introduction
The resourcer package is an R implementation of the data and computation resources description and connection. It resuses many existing R packages for reading various data formats and connecting to external data storage or computation servers. The resourcer package’s role is to interpret a resource description object to build the appropriate resource connection object. Because the bestiary of resources is very wide, the resourcer package provides a framework for dynamically extending the interpretation capabilities to new types of resources. This framework uses the object-oriented paradigm provided by the R6 library.
The purpose of the package is to allow access resources identified by a URL in a uniform way whether it references a dataset (stored in a file, a SQL table, a MongoDB collection etc.) or a computation unit (system commands, web services etc.). Usually some credentials will be defined, and an additional data format information can be provided to help dataset coercing to a data.frame object.
The main concepts are:
| Class | Description |
|---|---|
| resource | Access to a resource (dataset or computation unit) is described by an object with URL, optional credentials and optional data format properties. |
| ResourceResolver | A ResourceClient factory based on the URL scheme and available in a resolvers registry. |
| ResourceClient | Realizes the connection with the dataset or the computation unit described by a resource. |
| FileResourceGetter | Connect to a file described by a resource. |
| DBIResourceConnector | Establish a DBI connection. |
7.2 File Resources
These are resources describing a file. If the file is in a remote location, it must be downloaded before being read. The data format specification of the resource helps to find the appropriate file reader.
7.2.1 File Getter
The file locations supported by default are:
file, local file system,http(s), web address, basic authentication,gridfs, MongoDB file store,scp, file copy through SSH,opal, Opal file store.
This can be easily applied to other file locations by extending the FileResourceGetter class. An instance of the new file resource getter is to be registered so that the FileResourceResolver can operate as expected.
7.2.2 File Data Format
The data format specified within the resource object, helps with finding the appropriate file reader. Currently supported data formats are:
- the data formats that have a reader in tidyverse: readr (
csv,csv2,tsv,ssv,delim), haven (spss,sav,por,dta,stata,sas,xpt), readxl (excel,xls,xlsx). This can be easily applied to other data file formats by extending the FileResourceClient class. - the R data format that can be loaded in a child R environment from which object of interest will be retrieved.
Usage example that reads a local SPSS file:
# make a SPSS file resource
res <- resourcer::newResource(
name = "CNSIM1",
url = "file:///data/CNSIM1.sav",
format = "spss"
)
# coerce the csv file in the opal server to a data.frame
df <- as.data.frame(res)To support other file data format, extend the FileResourceClient class with the new data format reader implementation. The associated factory class, an extension of the ResourceResolver class, is also to be implemented and registered.
7.3 Database Resources
7.3.1 DBI Connectors
DBI is a set of virtual classes that are are used to abstract the SQL database connections and operations within R. Thus any DBI implementation can be used to access to a SQL table. Which DBI connector to be used is an information that can be extracted from the scheme part of the resource’s URL. For instance a resource URL starting with postgres:// will require the RPostgres driver. To separate the DBI connector instantiation from the DBI interface interactions in the SQLResourceClient, a DBIResourceConnector registry is to be populated. The currently supported SQL database connectors are:
mariadbMariaDB connector,mysqlMySQL connector,postgresorpostgresqlPostgres connector,presto,presto+httporpresto+httpsPresto connector,spark,spark+httporspark+httpsSpark connector.
To support another SQL database with a DBI driver, extend the DBIResourceConnector class and register it:
7.3.2 Use dplyr
Having the data stored in the database allows to handle large (common SQL databases) to big (PrestoDB, Spark) datasets using dplyr which will delegate as many operations as possible to the database.
7.3.3 Document Databases
NoSQL databases can be described by a resource. The nodbi can be used here. Currently only connection to MongoDB database is supported using URL scheme mongodb or mongodb+srv.
7.4 Computation Resources
Computation resources are resources on which tasks/commands can be triggered and from which resulting data can be retrieved.
Example of computation resource that connects to a server through SSH:
# make an application resource on a ssh server
res <- resourcer::newResource(
name = "supercomp1",
url = "ssh://server1.example.org/work/dir?exec=plink,ls",
identity = "sshaccountid",
secret = "sshaccountpwd"
)
# get ssh client from resource object
client <- resourcer::newResourceClient(res) # does a ssh::ssh_connect()
# execute commands
files <- client$exec("ls") # exec 'cd /work/dir && ls'
# release connection
client$close() # does ssh::ssh_disconnect(session)7.5 Interacting with R Resources
As the ResourceClient is just a connector to a resource, to make this useful some data conversion functions are defined by default:
- R data.frame, which is the most common representation of tabular data in R. A data frame, as defined in R base, is an object stored in memory that may be not suitable for large to big datasets.
- dplyr tbl, which is another representation of tabular data that nicely integrates with the DBI: filtering, mutation and aggregation operations can be delegated to the underlying SQL database, reducing the R memory and computation footprint. Useful functions are also provided to perform join operations on relational datasets. A data.frame can be accessed as a tbl and vice versa.
In the case when the resource is a R object, the RDataFileResourceClient offers the ability to get the internal raw data object. Then complex data structures, optimized for a specific research domain, can be accessed with the most appropriate tools.
When the resource is a computation service provider, the interaction with the resource client will consist of issuing commands/requests with parameters and getting the result from it either as a response object or as a file to be downloaded.
Another way of interacting with a resource is to get the internal connection object (a database connector, a SSH connector etc.) from the ResourceClient and then apply any kind of operations that are defined for it. The general purpose of a resource is not to substitute itself to the underlying library, it is to facilitate the access to the related data and services.
7.6 Extending Resources
There are several ways to extend the Resources handling. These are based on different R6 classes having a isFor(resource) function:
- If the resource is a file located at a place not already handled, write a new FileResourceGetter subclass and register an instance of it with the function
registerFileResourceGetter(). - If the resource is a SQL engine having a DBI connector defined, write a new DBIResourceConnector subclass and register an instance of it with the function
registerDBIResourceConnector(). - If the resource is in a domain specific web application or database, write a new ResourceResolver subclass and register an instance of it with the function
registerResourceResolver(). This ResourceResolver object will create the appropriate ResourceClient object that matches your needs.
The design of the URL that will describe your new resource should not overlap an existing one, otherwise the different registries will return the first instance for which the isFor(resource) is TRUE. In order to distinguish resource locations, the URL’s scheme can be extended, for instance the scheme for accessing a file in a Opal server is opal+https so that the credentials be applied as needed by Opal.
In order to simplify the usage of these resource client classes, the resourcer package combines several software design patterns:
- The factory pattern, a classic creational design pattern. It is realized by the ResourceResolver R6 class which is responsible for making a ResourceClient object matching a resource object provided.
- The registry pattern, “a well-known object that other objects can use to find common objects and services” as described by M. Fowler. It is basically a global list of objects to iterate through to find the appropriate one. In the resourcer package there are several registries: (1) the registry of ResourceResolver objects (the ResourceClient factories), (2) the registry of FileResourceGetter objects and (3) the registry of DBIResourceConnector objects.
- The self-registration pattern, which consists of delegating the registration of new services to their provider. The package event mechanism of R is used so that an R package self-registers its resource components in the registries previously mentioned when the package is loaded at runtime.
For implementing ResourceClient factories the resourcer package provides different ResourceResolver classes (see Figure 7.1):
- for file resources, which will discriminate the resources based on the URL property (checking if any FileResourceGetter exists for that resource) and the data format property (for getting additional information about how to read the data). The FileResourceGetter can be extended to new file locations: as an example, the s3.resourcer R package is able to get resource files from Amazon Web Service S3 file stores.
- for database resources, which will discriminate the resources based on the scheme part of the URL (checking if any DBIResourceConnector can be found or whether a nodbi connector can be created). The DBIResourceConnector can be extended to new DBI implementations. For instance, using bigrquery R package, it would be easy to implement access to a resource stored in a Google’s BigQuery database.
- for command-based computation resources, which will discriminate the resources based on the scheme part of the URL, indicating how to issue commands (local shell or secure remote shell).
Figure 7.1: ResourceResolver class diagram
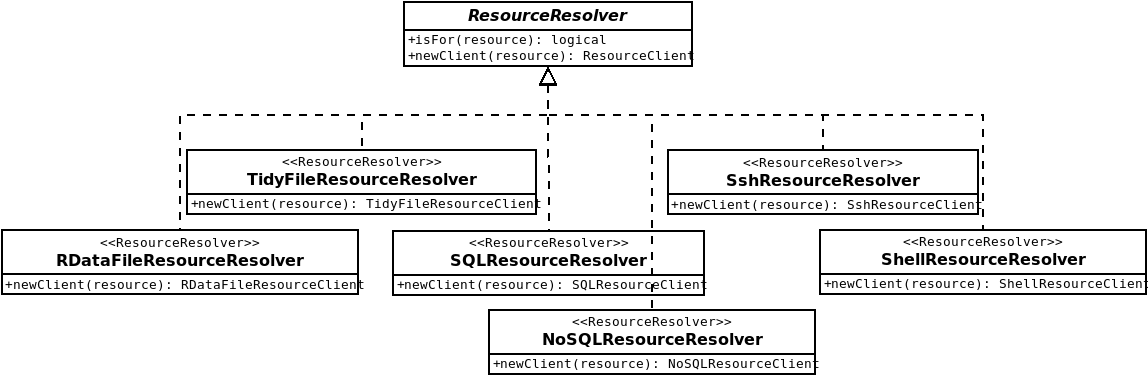
Additional ResourceResolver and ResourceClient extensions could be implemented for accessing domain specific applications which would expose data extraction and/or analysis services, the only requirement is that an R connection API exists for the considered application.
The process of creating a new ResourceClient instance from a resource object consists of iterating over a registry of ResourceResolver instances and finding the one that can handle the resource object by inspecting its properties (URL, data format etc.). The URL property inspection can imply a lookup in the additional registries: if the resource is a data file, the file resource resolvers will check in the FileResourceGetter registry whether there is one that applies; if the resource is a DBI-compatible database, the SQL resource resolver will check in the DBIResourceConnector registry whether there is one that applies. This workflow is simply triggered by a resourcer’s function call.
7.7 Resource Forms
As the definition a new resource can be error prone, when a URL is complex, or when there is a limited choice of formats or when credentials can be on different types, it is recommended to declare the resources forms and factory functions within the R package. This resource declaration is to be done in JavaScript, as this is a very commonly used language for building graphical user interfaces.
These files are expected to be installed at the root of the package folder (then in the source code of the R package, they will be declared in the inst/resources folder), so that an external application can lookup the packages statically having declared some resources.
The configuration file inst/resources/resource.js is a javascript file which contains an object with the properties:
settings, a JSON object that contains the description and the documentation of the web forms (based on the json-schema specification).asResource, a javascript function that will convert the data captured from one of the declared web forms into a data structure representing theresourceobject.
As an example (see also resourcer’s resource.js):
var myPackage = {
settings: {
"title": "MyPackage resources",
"description": "MyPackage resources are for etc.",
"web": "https://github.com/org/myPackage",
"categories": [
{
"name": "my-format",
"title": "My data format",
"description": "Data are files in my format, that will be read by myPackage etc."
}
],
"types": [
{
"name": "my-format-http",
"title": "My data format - HTTP",
"description": "Data are files in my format, that will be downloaded from a HTTP server etc.",
"tags": ["my-format", "http"],
"parameters": {},
"credentials": {}
}
]
},
asResource: function(type, name, params, credentials) {
// make a resource object from arguments, using type to drive
// what params/credentials properties are to be used
// a basic example of resource object:
return {
"name": name,
"url": params.url,
"format": params.format,
"identity": credentials.username,
"secret": credentials.password
};
}
}The specifications for the settings object is the following:
| Property | Type | Description |
|---|---|---|
| title | string |
The title of the set of resources. |
| description | string |
The description of the set of resources. |
| web | string |
A web link that describes the resources. |
| categories | array of object |
A list of category objects which are used to categorize the declared resources in terms of resource location, format, usage etc. |
| types | array of object |
A list of type objects which contains a description of the parameters and credentials forms for each type of resource. |
Where the category object is:
| Property | Type | Description |
|---|---|---|
| name | string |
The name of the category that will be applied to each resource type, must be unique. |
| title | string |
The title of the category. |
| description | string |
The description of the category. |
And the type object is:
| Property | Type | Description |
|---|---|---|
| name | string |
The identifying name of the resource, must be unique. |
| title | string |
The title of the resource. |
| description | string |
The description of the resource form. |
| tags | array of string |
The tag names that are applied to the resource form. |
| parameters | object |
The form that will be used to capture the parameters to build the url and the format properties of the resource (based on the json-schema specification). Some specific fields can be used: _package to capture the R package name or _packages to capture an array of R package names to be loaded prior to the resource assignment. |
| credentials | object |
The form that will be used to capture the access credentials to build the identity and the secret properties of the resource (based on the json-schema specification). |
The asResource function is a javascript function which signature is function(type, name, params, credentials) where:
type, the form name used to capture the resource parameters and credentials,name, the name to apply to the resource,params, the captured parameters,credentials, the captured credentials.
The name of the root object must follow the pattern: <R package> (note that any dots (.) in the R package name are to be replaced by underscores (_)).
A real example of how to create this file for the `{r Githubpkg(“isglobal-brge”, “dsOmics”)} package (described in this Section) can be found here.
7.8 Examples of Resources
The resourcer package provides some common resources, out of the box. Additional resource extensions are also available as separate R packages. The following table lists some known resource implementations.
| Resource | R Package | Description |
|---|---|---|
| Tidy data files | resourcer |
Data files that have a reader from the tidyverse ecosystem. |
| R data files | resourcer |
Any R objects stored in files in Rdata or RDS formats. |
| SQL databases | resourcer |
Main open source SQL databases vendors are supported: MySQL, MariaDB and PostgreSQL |
| Apache Spark | resourcer |
Big data analytics system, accessible as a SQL database, with also additional computation capabilities (machine leaning API). |
| SSH server | resourcer |
Command based remote computation service. |
| MongoDB | resourcer |
A NoSQL database. |
| HL7 FHIR resource | fhir.resourcer | Data extractions from HL7 FIHR compliant servers, the standard for clinical data management systems. |
| Dremio dataset | odbc.resourcer | Dremio is a data lake engine, tailored for integrating big data. Dremio is accessible as a SQL database. |
| GDS data files | dsOmics | Reads omics data stored in Genomic Data Structure (GDS) format or in a format that can be converted to GDS (such as VCF). |
Whenever a data access format or protocol is defined by a standard API specification, there is most likely an R implementation of this API and then a corresponding resourcer extension can be easily implemented. For instance developing a resourcer extension to access genomic data stored in a GA4GH system should be possible and would be very powerful.
7.9 Using Resources with DataSHIELD
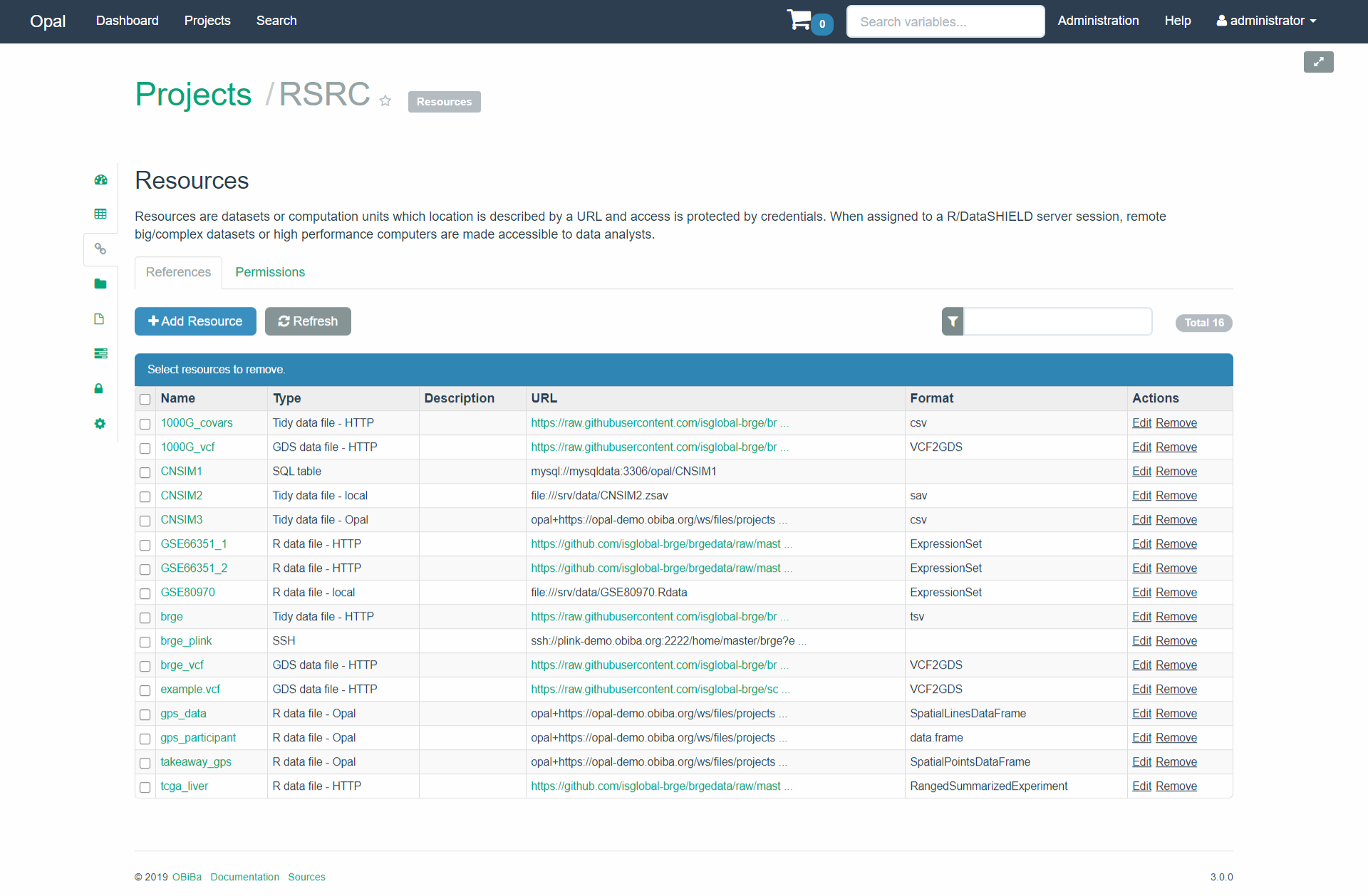
Let us illustrate how to deal with different types of resources within DataSHIELD. To this end, let use our Opal demo example (RSRC project) available at https://opal-demo.obiba.org which has the following resources
Figure 7.2: Resources from a demo enviroment available at https://opal-demo.obiba.org
7.9.1 TSV File into a tibble or data.frame
We start by illustrating how to get a simple TSV file (brge.txt) into the R server. This file is located at a GitHub repository: https://raw.githubusercontent.com/isglobal-brge/brgedata/master/inst/extdata/brge.txt and it is not necesary to be moved from there. This is one of the main strenght of the resources implementation.
This code describes how to get the resource (a TSV file) as a data.frame into the R Server. Note that this is secure access since a user name and password must be provided
library(DSOpal)
library(dsBaseClient)
# access to the 'brge' resource (NOTE: RSRC.brge is need since the project
# is called RSRC)
builder <- newDSLoginBuilder()
builder$append(server = "study1", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "password",
resource = "RSRC.brge", driver = "OpalDriver")
logindata <- builder$build()
# the resource is loaded into R as the object 'res'
conns <- datashield.login(logins = logindata, assign = TRUE,
symbol = "res")
# the resource is assigned to a data.frame
# Assign to the original R class (e.g ExpressionSet)
datashield.assign.expr(conns, symbol = "dat",
expr = quote(as.resource.data.frame(res)))
ds.class("dat")$study1
[1] "spec_tbl_df" "tbl_df" "tbl" "data.frame" 7.9.2 R Data File into a R object
Now let us describe how to get a specific type of R object into the R server. Our Opal demo contains a resource called GSE80970 which is located on a local machine (see Figure 7.2). The resource is an R object of class ExpressionSet which is normally used to jointly encapsulate gene expression, metadata and annotation. In general, we can retrieve any R object in its original format and if a method to coerce the specific object into a data.frame exists, we can also retrieve it as a tibble/data.frame.
# prepare login data and resource to assign
builder <- newDSLoginBuilder()
builder$append(server = "study1", url = "https://opal-demo.obiba.org",
user = "dsuser", password = "password",
resource = "RSRC.GSE80970", driver = "OpalDriver")
logindata <- builder$build()
# login and assign resource (to 'res' symbol)
conns <- datashield.login(logins = logindata, assign = TRUE,
symbol = "res")
# coerce ResourceClient objects to a data.frame called 'DF'
# NOTE: as.data.frame exists for `ExpressionSet` objects
datashield.assign.expr(conns, symbol = "DF",
expr = quote(as.resource.data.frame(res)))
ds.class("DF")$study1
[1] "data.frame"# we can also coerce ResourceClient objects to their original format.
# This will allow the analyses with specific R/Bioconductor packages
datashield.assign.expr(conns, symbol = "ES",
expr = quote(as.resource.object(res)))
ds.class("ES")$study1
[1] "ExpressionSet"
attr(,"package")
[1] "Biobase"