rexposome project
rexposome project is a set of R packages developed at Bioinformatics Research Group in Epidemiology (BRGE) from Barcelona Global Health Institute (ISGlobal). The project aims to provide a framework to incorporate exposome data in R/Bioconductor pipelines, with the goal of describing and analyzin exposome data.
Currently, the Bioconductor packages included in rexposome project are rexposome and omicRexposome. The first package, rexposome, offers an interface to load exposome data into Bioconductor-like objects and functions to describe and characterize the exposome. It also includes functions to perform Exposome-Wide Association Studies. The second package, omicRexposome, contains functions to perform exposome-omic association studies and multi-omic integration of exposome and omic data-sets.
Package Features
exposome data loading
rexposome implements readExposome and loadExposme functions. The first one is in charge to load into R exposome data stored in raw file and to create an ExposomeSet. The second one creates an ExposomeSet from already existing data.frames.
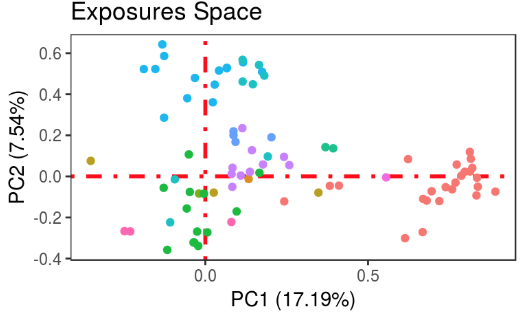
exposome data exploration
Several functions are implemented in rexposome to explore and describe exposome data. The method plotMissings allow to analyse the amount of missing data. plotHistogram and plotFamily allow to see the behaviour of thee exposures. correlation and plotCorrelation helps to understand the relation between exposures.
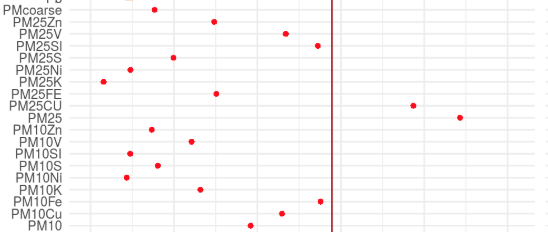
exposome data analysis
rexposome allows to perform EXposome-Wide Association Study (EXWAS) in both univariate and multivariate ways using standard single-exposure association test or through an elastic net approach.
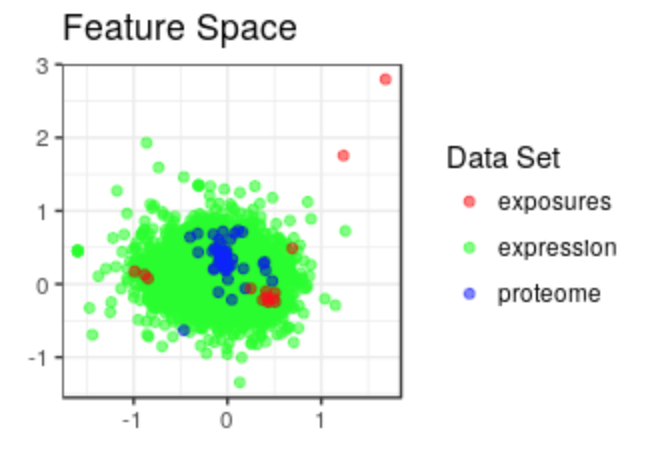
exposome-omic data analysis
omicRexposome implement the functions associations and integration. The fist allows to test the association between exposures and omic features (genes, CpGs...). The second allows to perform an integration of multiple layers, including exposome data.