3 Data sets
3.1 Exposome dataset
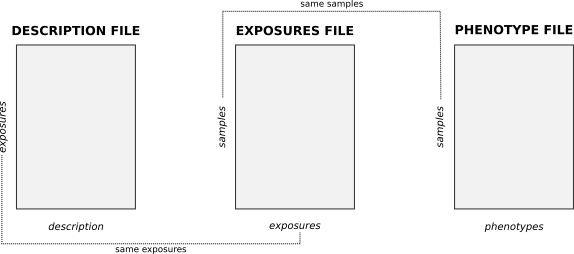
The exposome is composed of three different files (in *.csv, *.tsv or *.txt format). Those files are refered inside the Shiny as exposures, description and phenotypes. Their content is the following:
- The
exposuresfile contains the measures of each exposure for all the individuals included on the analysis. It is a matrix-like file having a row per individual and a column per exposures. It must includes a column with the subject’s identifier. - The
descriptionfile contains a row for each exposure and, at last, defined the families of exposures. Usually, this file incorporates a description of the exposures, the matrix where it was obtained and the units of measurement among others. - The
phenotypesfile contains the covariates to be included in the analysis as well as the health outcomes of interest. It contains a row per individual included in the analysis and a column for each covariate and outcome. Moreover, it must include a column with the individual’s identifier.
Some remarks regarding this files:
- All three files have to share the same separator element, for
*.csvfiles is typical to use a comma (,) but it could also be a semicolon (;). - The exposure names have to start with a character [a-z/A-Z], leading special characters will cause the data entry to return errors.
- Exactly the same exposures have to be present on the description and exposures files.
- Exactly the same samples have to be present on the exposures and phenotypes files.
- The exposures and phenotypes files have an ID column, the description file does not have an ID column nor row.
A visual representation of the three matrices and how they correlate is the following.
Exposures data file example:
id bde100 bde138 bde209 PFOA ...
sub01 2.4665 0.7702 1.6866 2.0075 ...
sub02 0.7799 1.4147 1.2907 1.0153 ...
sub03 -1.6583 -0.9851 -0.8902 -0.0806 ...
sub04 -1.0812 -0.6639 -0.2988 -0.4268 ...
sub05 -0.2842 -0.1518 -1.5291 -0.7365 ...
... ... ... ... ...Description data file example:
exposure family matrix description
bde100 PBDEs colostrum BDE 100 - log10
bde138 PBDEs colostrum BDE 138 - log10
bde209 PBDEs colostrum BDE 209 - log10
PFOA PFAS cord blood PFOA - log10
PFNA PFAS cord blood PFNA - log10
PFOA PFAS maternal serum PFOA - log10
PFNA PFAS maternal serum PFNA - log10
hg Metals cord blood hg - log 10
Co Metals urine Co (creatinine) - log10
Zn Metals urine Zn (creatinine) - log10
Pb Metals urine Pb (creatinine) - log10
THM Water --- Average total THM uptake - log10
CHCL3 Water --- Average Chloroform uptake - log10
BROM Water --- Average Brominated THM uptake - log10
NO2 Air --- NO2 levels whole pregnancy- log10
Ben Air --- Benzene levels whole pregnancy- log10Phenotypes data file example:
id asthma BMI sex age ...
sub01 control 23.2539 boy 4 ...
sub02 asthma 24.4498 girl 5 ...
sub03 asthma 15.2356 boy 4 ...
sub04 control 25.1387 girl 4 ...
sub05 control 22.0477 boy 5 ...
... ... ... ... ...3.2 Plain datasets
If the researcher has gathered all the data on a single file which contains both phenotype and exposure data, this file can be used too. The user interface has a selector for it, more information on the correspondent section.
A visual representation of a plain dataset is the following.

Plain dataset example (3 exposures + 2 phenotypes):
id bde100 bde138 bde209 asthma BMI ...
sub01 2.4665 0.7702 1.6866 control 23.2539 ...
sub02 0.7799 1.4147 1.2907 asthma 24.4498 ...
sub03 -1.6583 -0.9851 -0.8902 asthma 15.2356 ...
sub04 -1.0812 -0.6639 -0.2988 control 25.1387 ...
sub05 -0.2842 -0.1518 -1.5291 control 22.0477 ...
... ... ... ... ... ...3.3 Omics dataset
The omics data inputed to the Shiny must be provided as an *.RData. This file has to contain an ExpressionSet, which is an S4 object. This object is a data container of the Bioconductor toolset.
For further information on ExpressionSet and how to create and manipulate them, please visit the official documentation and this selected vignette.