OMOP DB Filter Population
A class for managing a test database with person, observation, and measurement data.
1"""
2A class for managing a test database with person, observation, and measurement data.
3"""
4from biomodel.database import OMOP_DB
5from biomodel.models import Framingham
6from biomodel.biomodel import OMOPId
7from pathlib import Path
8
9if __name__ == '__main__':
10
11 data_path = Path("/home/josalhor/Desktop/bioTwin/data/ukbiobank-simulation")
12
13
14 class TestDB(OMOP_DB):
15 """
16 A class for managing a test database with person, observation, and measurement data.
17 """
18
19 person_path = data_path / "person.csv"
20 observation_path = data_path / "observation.csv"
21 measurement_path = data_path / "measurement.csv"
22
23
24 coltot = OMOPId(4224820)
25 hdl = OMOPId(4011133)
26 tas = OMOPId(4152194)
27 tad = OMOPId(4154790)
28 smoke = OMOPId(1585856)
29 diab = OMOPId(35817874)
30
31 db = TestDB()
32 model = Framingham()
33 woman = 2
34 man = 1
35
36
37 def filter(params):
38 """
39 Filters the input parameters based on the specified conditions.
40
41 Args:
42 params (dict): A dictionary containing input parameters.
43
44 Returns:
45 bool: True if the input parameters meet the specified conditions, False otherwise.
46
47 """
48 # age = params[OMOPId("age")]
49 # return age > 35 and age < 70
50 sex = params[OMOPId("sex")]
51 return sex == man
52
53
54 prediction = model.predict_population_time(
55 db,
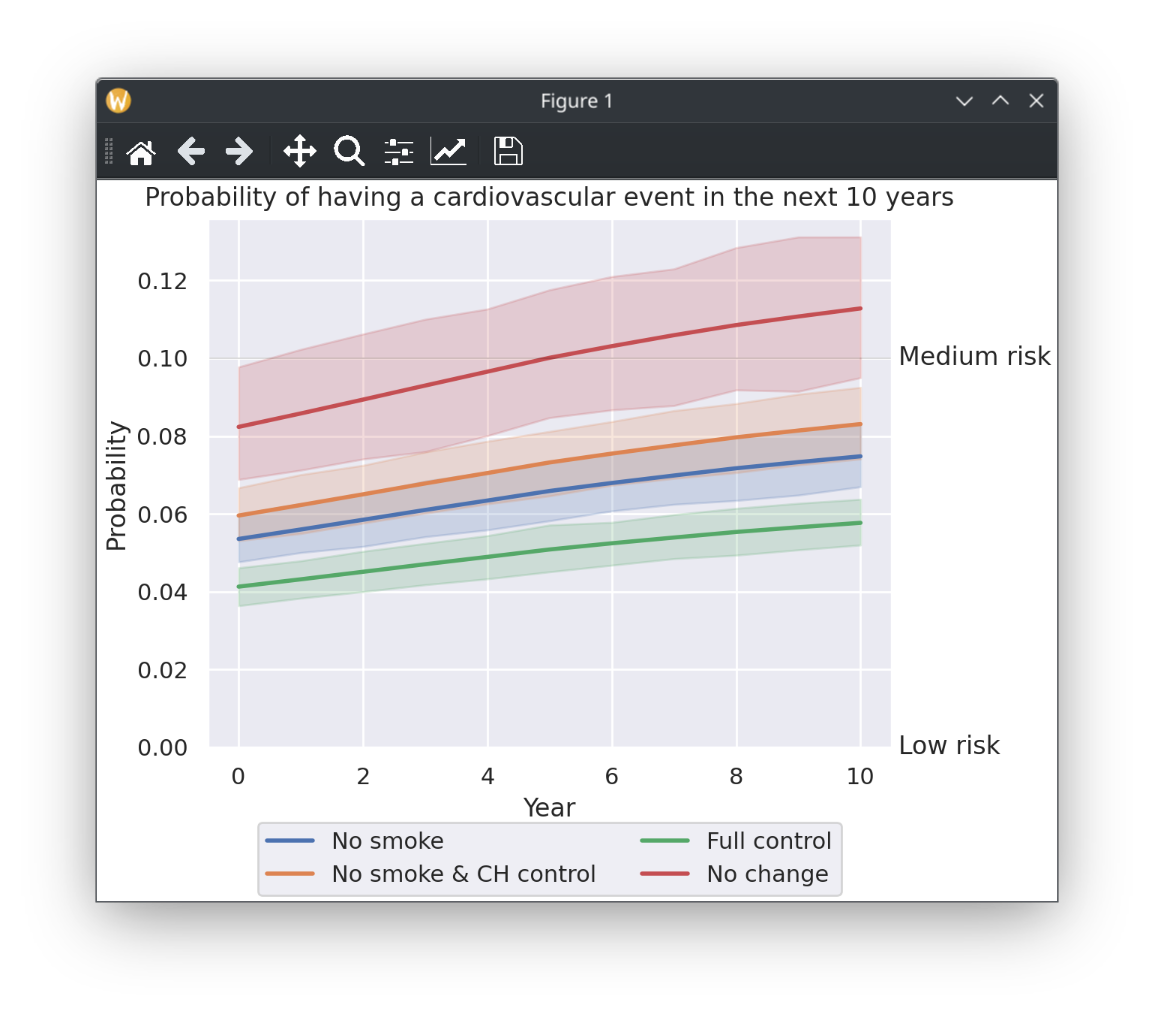
56 {
57 "No smoke": {smoke: 0},
58 "No smoke & CH control": {smoke: 0, coltot: 180, hdl: 50},
59 "Full control": {
60 coltot: 180,
61 hdl: 50,
62 tas: 125,
63 tad: 80,
64 smoke: 0,
65 },
66 },
67 person_filter=filter,
68 )
69
70 prediction.show_change()