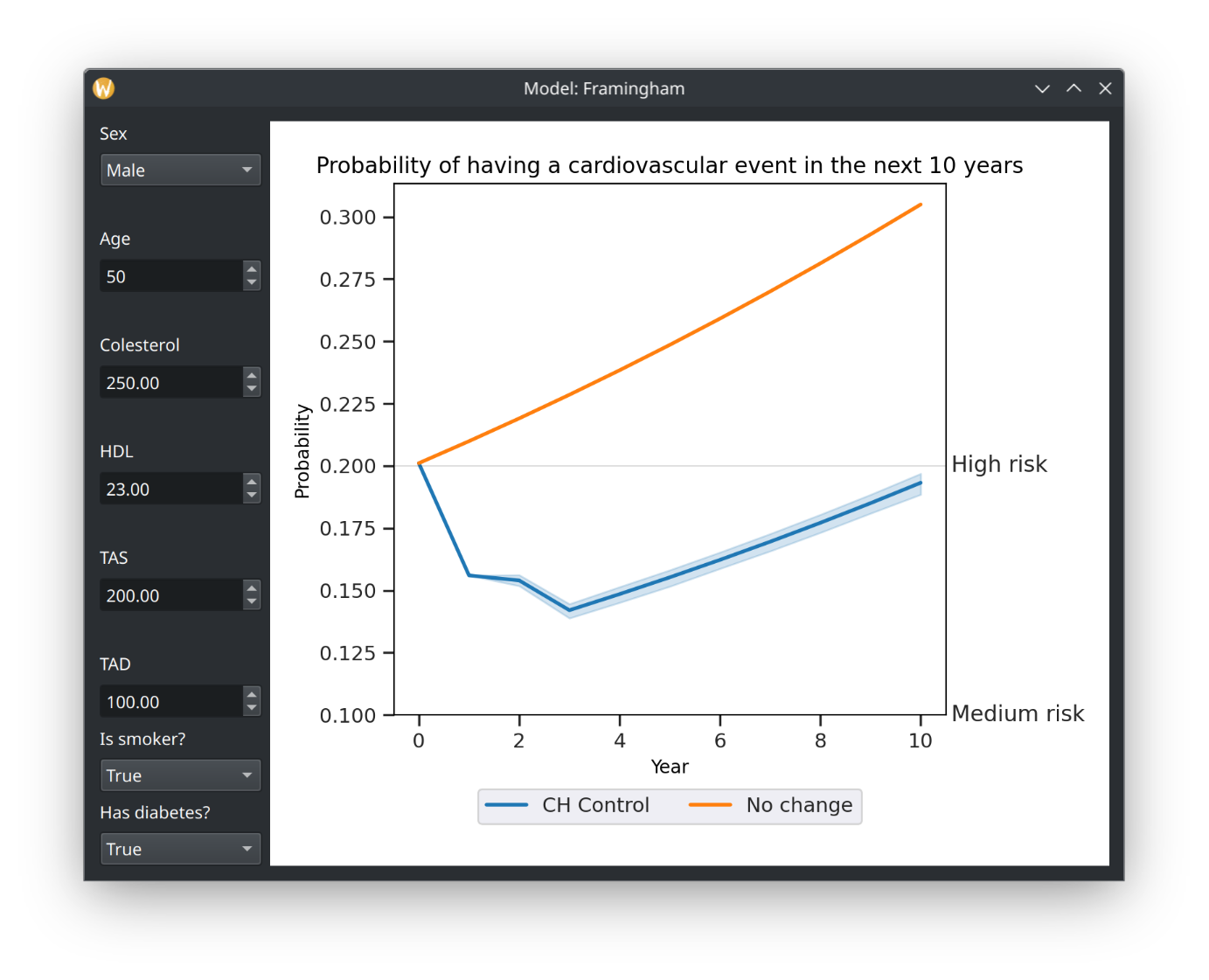
Framingham GUI Simulation with Normal Distribution
This file contains code for running the Framingham model using biomodel. It defines input variables such as sex, age, cholesterol levels, blood pressure, smoking status, and diabetes status. It also includes a variation for cholesterol control. The code then initializes the model, provides input data, applies the variation, and runs the model.
1"""
2This file contains code for running the Framingham model using biomodel. It defines input variables such as sex, age, cholesterol levels, blood pressure, smoking status, and diabetes status. It also includes a variation for cholesterol control. The code then initializes the model, provides input data, applies the variation, and runs the model.
3"""
4from biomodel.models import Framingham
5from biomodel.gui import InterfaceModel
6from biomodel.biomodel import OMOPId
7from biomodel.transformations import Before, Normal, Reduce
8
9if __name__ == '__main__':
10
11 sex = OMOPId("sex")
12 age = OMOPId("age")
13 coltot = OMOPId(4224820)
14 hdl = OMOPId(4011133)
15 tas = OMOPId(4152194)
16 tad = OMOPId(4154790)
17 smoke = OMOPId(1585856)
18 diab = OMOPId(35817874)
19
20 current = {
21 sex: 1,
22 age: 50,
23 coltot: 250,
24 hdl: 23,
25 tas: 200,
26 tad: 100,
27 smoke: 1,
28 diab: 1,
29 }
30
31 variation = {
32 "CH Control": [
33 Before(year=4, transformation=Reduce(id=coltot, value=Normal(mean=24, std=6)))
34 ],
35 }
36 model = InterfaceModel(Framingham(), current, variation)
37 model.run()